3APW
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APU
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein | | Descriptor: | Alpha-1-acid glycoprotein 2, TETRAETHYLENE GLYCOL | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
8Z75
 
 | | The structure of non-activated thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-04-19 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of non-activated thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH)
To Be Published
|
|
8Z76
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 during 6 months | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-04-19 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2
To Be Published
|
|
8JBN
 
 | | Vascular endothelial protein tyrosine phosphatase in complex with Cpd-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(1~{H}-indol-3-yl)-1,2-oxazole-3-carboxylic acid, ... | | Authors: | Orita, T, Furuzono, T, Doi, S, Adachi, T. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Novel VE-PTP Inhibitors Using Orthogonal Biophysical Techniques.
Biochemistry, 62, 2023
|
|
8JBY
 
 | | Vascular endothelial protein tyrosine phosphatase in complex with Cpd-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(hydroxymethyl)-5-(1-methylindol-3-yl)-1,2-oxazole-3-carboxylic acid, ... | | Authors: | Orita, T, Furuzono, T, Doi, S, Adachi, T. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Novel VE-PTP Inhibitors Using Orthogonal Biophysical Techniques.
Biochemistry, 62, 2023
|
|
8YTR
 
 | | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, CopC domain-containing protein, DI(HYDROXYETHYL)ETHER | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Nikolaeva, A.Y, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YTS
 
 | | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution | | Descriptor: | Cytochrome C, HEME C | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution
To Be Published
|
|
8YU6
 
 | | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate
To Be Published
|
|
8YOU
 
 | | The pmTcDH complex structure with an inhibitor SeCN | | Descriptor: | COPPER (II) ION, GLYCEROL, SELENIUM ATOM, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The pmTcDH complex structure with an inhibitor SeCN
To Be Published
|
|
8YTQ
 
 | | The structure of apoCopC from Thioalkalivibrio paradoxus | | Descriptor: | ACETATE ION, COPPER (II) ION, CopC domain-containing protein, ... | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of apoCopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YU5
 
 | | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q)
To Be Published
|
|
8Z77
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 and Na ascorbate during 12 hours | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Minyaev, M.E, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-04-19 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 and Na ascorbate during 12 hours
To Be Published
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
4WVA
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4WVC
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4WVB
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
8RV0
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite | | Descriptor: | CALCIUM ION, HEME C, NITRIC OXIDE, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-01-31 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite
To Be Published
|
|
8RXU
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite reductase, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21
To Be Published
|
|
8RVM
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite c cytochrome c reductase, ... | | Authors: | Polyakov, K.M, Safoonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63
To Be Published
|
|
4XPO
 
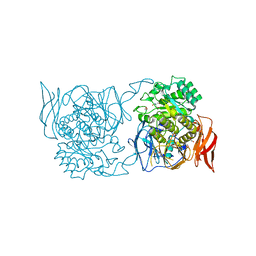 | | Crystal structure of a novel alpha-galactosidase from Pedobacter saltans | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPR
 
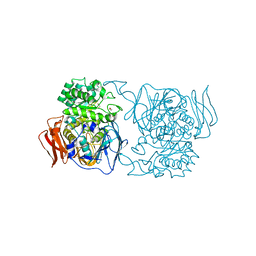 | | Crystal structure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPP
 
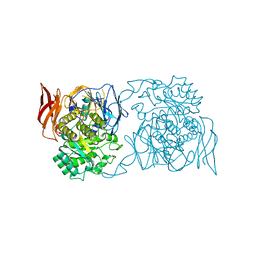 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
