6O2S
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2Q
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5ARD
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5J8X
 
 | | CRYSTAL STRUCTURE OF E. COLI PBP5 WITH 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, D-alanyl-D-alanine carboxypeptidase DacA | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
4EO4
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with seryl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonine--tRNA ligase, mitochondrial, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Soll, D, Simonovic, M. | | Deposit date: | 2012-04-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase.
J.Biol.Chem., 287, 2012
|
|
6ZTA
 
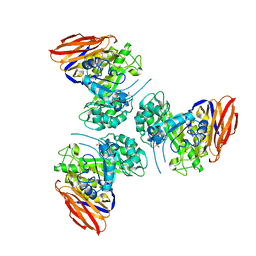 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6S0I
 
 | |
6Y6J
 
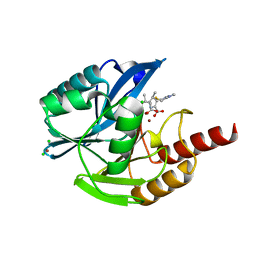 | | VIM-2 in Complex with Biapenem Imine and Enamine Hydrolysis Products | | Descriptor: | Biapenem Enamine hydrolysis product, Biapenem Imine hydrolysis product, CHLORIDE ION, ... | | Authors: | Lucic, A, Schofield, C.J, Brem, J, McDonough, M.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of metallo-beta-lactamase VIM-2 in complex with the imine and enamine form of hydrolysed Biapenem
To Be Published
|
|
7BCS
 
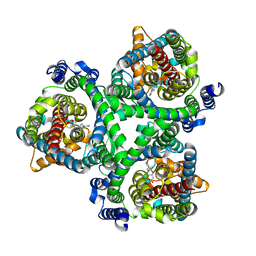 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "down") in the outward-open conformation. | | Descriptor: | (2~{S},4~{S})-4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7P55
 
 | |
7P5Q
 
 | |
7P5S
 
 | |
7P5G
 
 | |
4ZDO
 
 | |
7E8Z
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with SS81 | | Descriptor: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Tam, N.Y, Ng, Y.M, Shishodia, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
5APA
 
 | | Crystal structure of human aspartate beta-hydroxylase isoform a | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ASPARTYL/ASPARAGINYL BETA-HYDROXYLASE, ... | | Authors: | Krojer, T, Kochan, G, Pfeffer, I, McDonough, M.A, Pilka, E, Hozjan, V, Allerston, C, Muniz, J.R, Chaikuad, A, Gileadi, O, Kavanagh, K, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2015-09-15 | | Release date: | 2015-09-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
7BCQ
 
 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCT
 
 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DQR
 
 | | Crystal structure of Sulfurisphaera tokodaii methylated O6-methylguanine methyltransferase | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsonoda, M. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8CKF
 
 | | Crystal Structure of the first bromodomain of human BRD4 L94C variant in complex with racemic 3,5-dimethylisoxazol ligand | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{S})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Thomas, A.M, McDonough, M.A, Schiedel, M, Conway, S.J. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutate and Conjugate: A Method to Enable Rapid In-Cell Target Validation.
Acs Chem.Biol., 18, 2023
|
|
7EUV
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Hoshina, M, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with Flurbiprofen
To Be Published
|
|
3NJY
 
 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
6I30
 
 | | Crystal structure of the AmpC from Pseudomonas aeruginosa with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Class C beta-lactamase PDC-301 | | Authors: | Brem, J, Cahill, S.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2018-11-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the inhibition of AmpC and other beta-lactamases by cyclic boronates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4BU2
 
 | | 60S ribosomal protein L27A histidine hydroxylase (MINA53) in complex with Ni(II) and 2-oxoglutarate (2OG) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE MINA, ... | | Authors: | Chowdhury, R, Clifton, I.J, McDonough, M.A, Ng, S.S, Pilka, E, Oppermann, U, Schofield, C.J. | | Deposit date: | 2013-06-19 | | Release date: | 2014-05-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
