5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
3LNQ
 
 | | Structure of Aristaless homeodomain in complex with DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*TP*TP*AP*AP*AP*CP*CP*C)-3', 5'-D(*GP*GP*GP*TP*TP*TP*AP*AP*TP*TP*AP*GP*GP*G)-3', ACETATE ION, ... | | Authors: | Takamura, Y, Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
4YSD
 
 | | Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | ACETIC ACID, COPPER (II) ION, Nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSQ
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.38 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSP
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSA
 
 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSO
 
 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSS
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.7 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSU
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5D4I
 
 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KNC
 
 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KNB
 
 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5F7A
 
 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7B
 
 | | Resting state structure of CuNiR form Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6GF0
 
 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | Descriptor: | Lysozyme C | | Authors: | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5IFF
 
 | | Crystal structure of R.PabI-nonspecific DNA complex | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*AP*GP*TP*TP*CP*GP*AP*AP*CP*TP*AP*GP*TP*GP*C)-3'), Uncharacterized protein | | Authors: | Wang, D, Miyazono, K, Tanokura, M. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the restriction DNA glycosylase R.PabI in complex with nonspecific double-stranded DNA.
Sci Rep, 6, 2016
|
|
5NDC
 
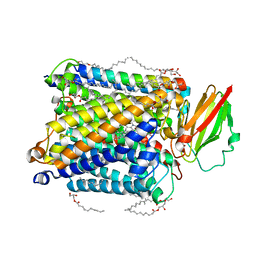 | | Structure of ba3-type cytochrome c oxidase from Thermus thermophilus by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide IIA, ... | | Authors: | Andersson, R, Safari, C, Dods, R, Nango, E, Tanaka, R, Nakane, T, Tono, K, Joti, Y, Bath, P, Dunevall, E, Bosman, E, Nureki, O, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature.
Sci Rep, 7, 2017
|
|
