1UHM
 
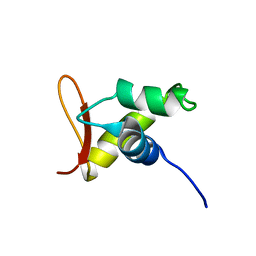 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | Descriptor: | Histone H1 | | Authors: | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-05 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
5XOC
 
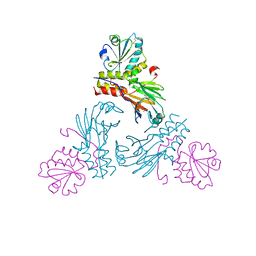 | | Crystal structure of human Smad3-FoxH1 complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Thioredoxin 1,Forkhead box protein H1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
4YOW
 
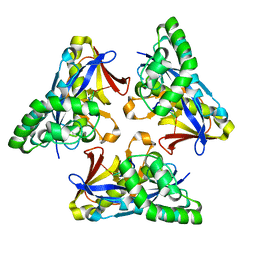 | |
4YOX
 
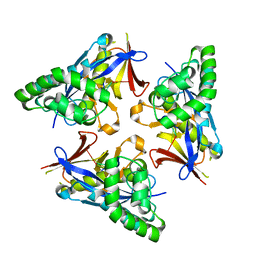 | |
4YOU
 
 | |
4YOR
 
 | |
4YOY
 
 | |
4YOV
 
 | |
6M64
 
 | | Crystal structure of SMAD2 in complex with CBP | | Descriptor: | CBP, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Ito, T, Wada, H, Tanokura, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
7CFA
 
 | |
7CO1
 
 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
8EQ1
 
 | |
8EU4
 
 | |
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
8EDT
 
 | |
8EDR
 
 | |
8EDS
 
 | |
8EQ0
 
 | |
8EQ3
 
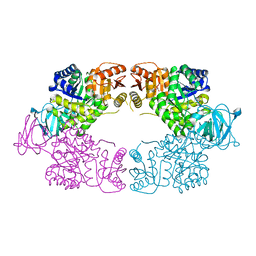 | | Escherichia coli pyruvate kinase A301T | | Descriptor: | IMIDAZOLE, MALONATE ION, Pyruvate kinase, ... | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-10-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
3JRS
 
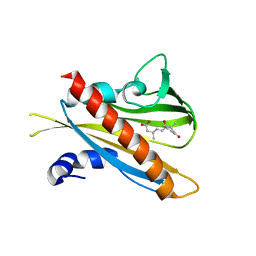 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
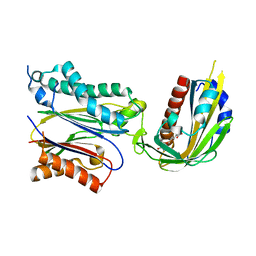 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
5XOD
 
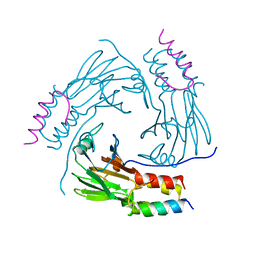 | | Crystal structure of human Smad2-Ski complex | | Descriptor: | Mothers against decapentaplegic homolog 2, Ski oncogene | | Authors: | Miyazono, K, Moriwaki, S, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
4YOT
 
 | |
1V3Z
 
 | |
4YNG
 
 | | Twinned pyruvate kinase from E. coli in the T-state | | Descriptor: | Pyruvate kinase I, SULFATE ION | | Authors: | Donovan, K.A, Dobson, R.C.J. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Grappling with anisotropic data, pseudo-merohedral twinning and pseudo-translational noncrystallographic symmetry: a case study involving pyruvate kinase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
