6UYO
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYX
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYU
 
 | | Crystal structure of K45-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYQ
 
 | | Crystal structure of K45-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYR
 
 | | Crystal structure of K46-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYY
 
 | | Crystal structure of K39-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6V7Q
 
 | | Crystal structure of SUMO1 in complex with phosphorylated PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6V7S
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6UYP
 
 | | Crystal structure of K39-acetylated SUMO1 in complex with PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYT
 
 | | Crystal structure of K39-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYZ
 
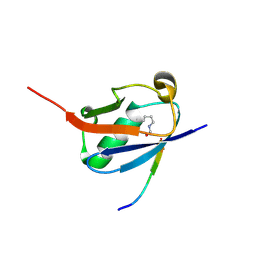 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYV
 
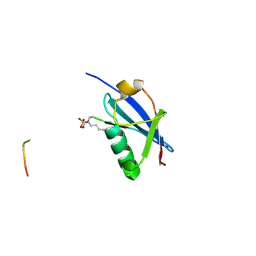 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6V7R
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6UYS
 
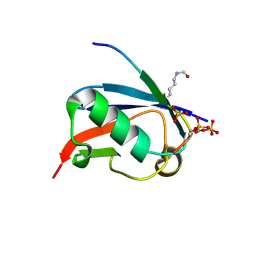 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6V7P
 
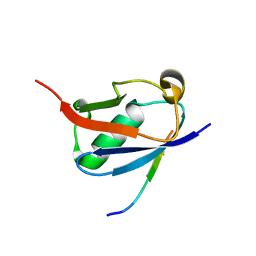 | | Crystal structure of SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
1NCP
 
 | |
1NHA
 
 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
1OLG
 
 | |
1OLH
 
 | |
1ONV
 
 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5U7A
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | Alkylmercury lyase, BROMIDE ION, Dimethyltin dibromide, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5SXP
 
 | | STRUCTURAL BASIS FOR THE INTERACTION BETWEEN ITCH PRR AND BETA-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Rho guanine nucleotide exchange factor 7 | | Authors: | Cappadocia, L, Desrochers, G, Lussier-Price, M, Angers, A, Omichinski, J.G. | | Deposit date: | 2016-08-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of interactions between SH3 domain-containing proteins and the proline-rich region of the ubiquitin ligase Itch.
J. Biol. Chem., 292, 2017
|
|
5U88
 
 | | Crystal structure of a MerB-triimethyllead complex. | | Descriptor: | ACETATE ION, Alkylmercury lyase, Trimethyllead bromide | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-11 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7B
 
 | | Crystal structure of a the tin-bound form of MerB formed from Diethyltin. | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U79
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
