5FQ5
 
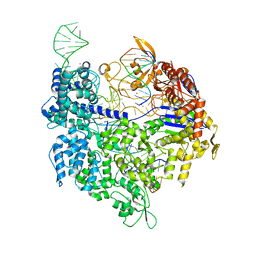 | | Crystal structure of Cas9-sgRNA-DNA complex solved by native SAD phasing | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Olieric, V, Weinert, T, Finke, A, Anders, C, Jinek, M, Wang, M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Data-Collection Strategy for Challenging Native Sad Phasing.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3DW4
 
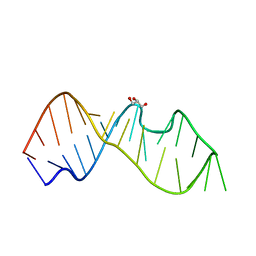 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DVZ
 
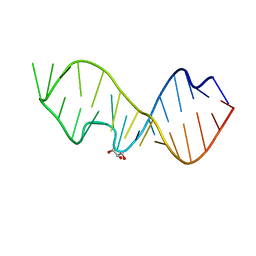 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW7
 
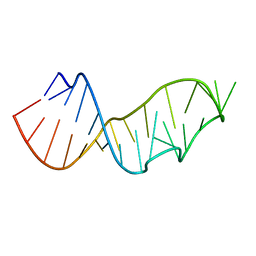 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
4TSZ
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TR7
 
 | |
4TR8
 
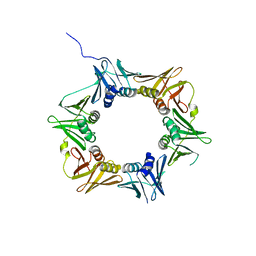 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
6QBY
 
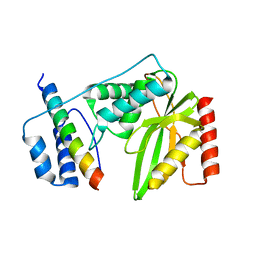 | | Crystal structure of VASH 2 in complex with SVBP | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Choi, S.R, Olieric, V, Steinmetz, M.O, Olieric, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6I59
 
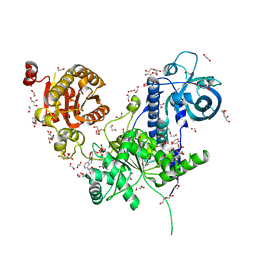 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6I5C
 
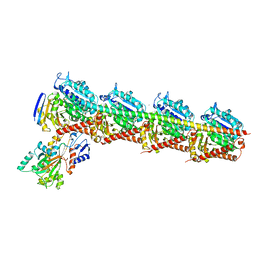 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
2WKK
 
 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | Descriptor: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | Deposit date: | 2009-06-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
1OK7
 
 | | A Conserved protein binding-site on Bacterial Sliding Clamps | | Descriptor: | DNA POLYMERASE III, DNA POLYMERASE IV | | Authors: | Burnouf, D.Y, Olieric, V, Wagner, J, Fujii, S, Reinbolt, J, Fuchs, R.P.P, Dumas, P. | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of Sliding Clamp/Ligand Interactions Suggest a Competition between Replicative and Translesion DNA Polymerases
J.Mol.Biol., 335, 2004
|
|
7DHG
 
 | | Crystal structure of SARS-CoV-2 Orf9b complex with human TOM70 | | Descriptor: | Mitochondrial import receptor subunit TOM70, ORF9b protein | | Authors: | Gao, X, Zhu, K, Qin, B, Olieric, V, Wang, M, Cui, S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions.
Nat Commun, 12, 2021
|
|
5OON
 
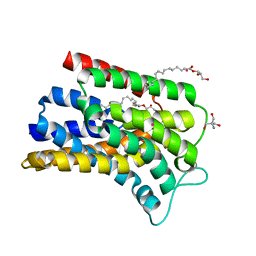 | | Structure of Undecaprenyl-Pyrophosphate Phosphatase, BacA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Wang, M, Howe, N, Ghachi, M.E.I, Weichert, D, Kerff, F, Stansfeld, P, Touze, T, Caffrey, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.
Nat Commun, 9, 2018
|
|
5OVJ
 
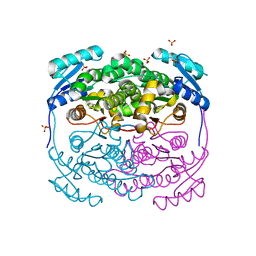 | | Structure of the apo form of Mycobacterium smegmatis MabA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OVK
 
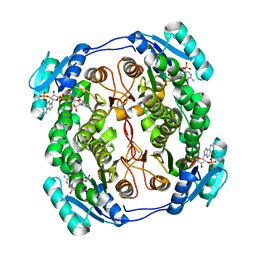 | | Crystal structure MabA bound to NADPH from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OVL
 
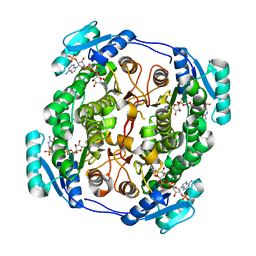 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
9FX7
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX6
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
3Q0X
 
 | | N-terminal coiled-coil dimer domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
3Q0Y
 
 | | N-terminal domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
