2E37
 
 | |
2EKM
 
 | |
3VGE
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGB
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGU
 
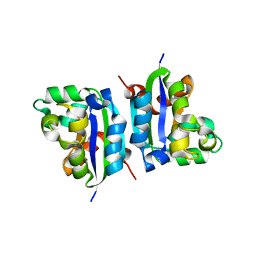 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGD
 
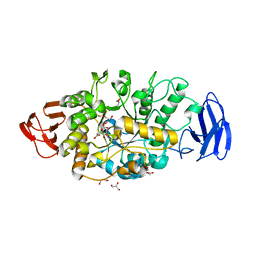 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGF
 
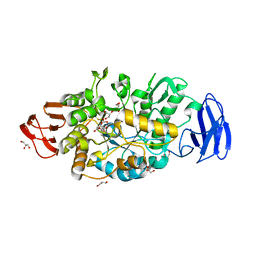 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGV
 
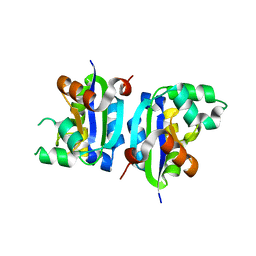 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGH
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGS
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGG
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGT
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
7XTL
 
 | |
7XTN
 
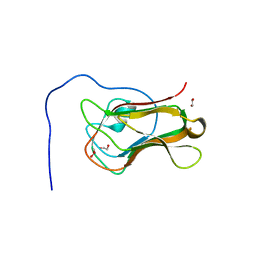 | | Crystal structure of the C-terminal domain of Bombyx mori N-acetylglucosaminyltransferase IV in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminyltransferase IV | | Authors: | Miyazaki, T, Oka, N, Mori, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure and sugar-binding ability of the C-terminal domain of N-acetylglucosaminyltransferase IV establish a new carbohydrate-binding module family.
Glycobiology, 32, 2022
|
|
7XTM
 
 | | Crystal structure of the C-terminal domain of Bombyx mori N-acetylglucosaminyltransferase IV | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, N-acetylglucosaminyltransferase IV | | Authors: | Miyazaki, T, Oka, N, Mori, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and sugar-binding ability of the C-terminal domain of N-acetylglucosaminyltransferase IV establish a new carbohydrate-binding module family.
Glycobiology, 32, 2022
|
|
6JV9
 
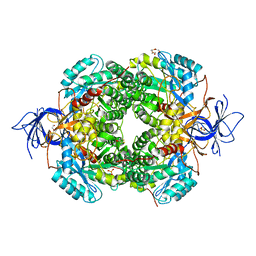 | | Crystal Structure of Human CRMP2 1-532, unmodified | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydropyrimidinase-related protein 2, ... | | Authors: | Jiang, X, Ogawa, T, Hirokawa, N. | | Deposit date: | 2019-04-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Enhanced carbonyl stress induces irreversible multimerization of CRMP2 in schizophrenia pathogenesis.
Life Sci Alliance, 2, 2019
|
|
3J7I
 
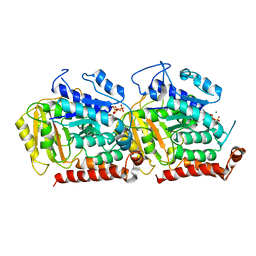 | | Structure of alpha- and beta- tubulin in GMPCPP-microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tubulin alpha-1A chain, ... | | Authors: | Yajima, H, Ogura, T, Nitta, R, Okada, Y, Sato, C, Hirokawa, N. | | Deposit date: | 2014-07-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy
J.Cell Biol., 198, 2012
|
|
3J6H
 
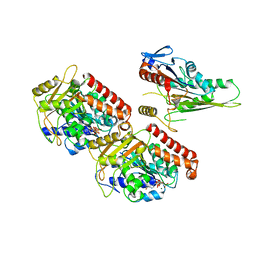 | | Nucleotide-free Kinesin motor domain complexed with GMPCPP-microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin heavy chain isoform 5C, MAGNESIUM ION, ... | | Authors: | Morikawa, M, Yajima, H, Nitta, R, Inoue, S, Ogura, T, Sato, C, Hirokawa, N. | | Deposit date: | 2014-02-21 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | X-ray and Cryo-EM structures reveal mutual conformational changes of Kinesin and GTP-state microtubules upon binding
Embo J., 2015
|
|
1IA0
 
 | | KIF1A HEAD-MICROTUBULE COMPLEX STRUCTURE IN ATP-FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-22 | | Release date: | 2002-03-22 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Switch-based Mechanism of Kinesin Motors
Nature, 411, 2001
|
|
5GSY
 
 | | Kinesin-8 motor, KIF19A, in the nucleotide-free state complexed with GDP-taxol microtubule | | Descriptor: | Kinesin-like protein KIF19 | | Authors: | Morikawa, M, Nitta, R, Yajima, H, Shigematsu, H, Kikkawa, M, Hirokawa, N. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
1I6I
 
 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-AMPPCP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
5GSZ
 
 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | Authors: | Wang, D, Nitta, R, Hirokawa, N. | | Deposit date: | 2016-08-18 | | Release date: | 2016-09-28 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
1I5S
 
 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-02-28 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1UC4
 
 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
1V8J
 
 | | The Crystal Structure of the Minimal Functional Domain of the Microtubule Destabilizer KIF2C Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION | | Authors: | Ogawa, T, Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.
Cell(Cambridge,Mass.), 116, 2004
|
|
