9JST
 
 | |
9JSX
 
 | |
9JSV
 
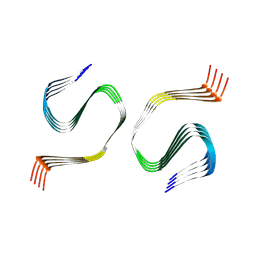 | | G175S mutant native PMEL amyloid | | Descriptor: | M-alpha | | Authors: | Oda, T, Yanagisawa, H. | | Deposit date: | 2024-10-01 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (1.79 Å) | | Cite: | Cryo-EM of PMEL Amyloids Reveals Pathogenic Mechanism of G175S in Pigment Dispersion Syndrome.
To Be Published
|
|
9JSW
 
 | |
9JSU
 
 | |
7WZ8
 
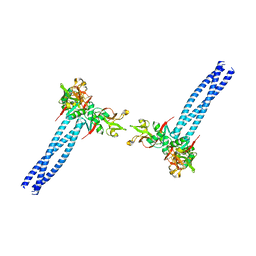 | | Structure of human langerin complex in Birbeck granules | | Descriptor: | SNAP-tag,C-type lectin domain family 4 member K | | Authors: | Oda, T, Yanagisawa, H. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-electron tomography of Birbeck granules reveals the molecular mechanism of langerin lattice formation.
Elife, 11, 2022
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
8JJ5
 
 | | Porcine uroplakin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tetraspanin, ... | | Authors: | Oda, T, Yanagisawa, H, Kikkawa, M. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane.
Commun Biol, 6, 2023
|
|
5H6J
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6L
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
6KLT
 
 | | Troponin of cardiac thin filament in low-calcium state | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Oda, T, Yanagisawa, H, Wakabayashi, T. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin.
J.Struct.Biol., 209, 2020
|
|
6KLU
 
 | | Troponin of cardiac thin filament in high-calcium state | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Oda, T, Yanagisawa, H, Wakabayashi, T. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin.
J.Struct.Biol., 209, 2020
|
|
6KLQ
 
 | |
6KLP
 
 | |
6KLL
 
 | | F-actin of cardiac thin filament in low-calcium state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oda, T, Yanagisawa, H, Wakabayashi, T. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin.
J.Struct.Biol., 209, 2020
|
|
6KLN
 
 | | F-actin of cardiac thin filament in high-calcium state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oda, T, Yanagisawa, H, Wakabayashi, T. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin.
J.Struct.Biol., 209, 2020
|
|
3A8R
 
 | | The structure of the N-terminal regulatory domain of a plant NADPH oxidase | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Oda, T, Hashimoto, H, Kuwabara, N, Akashi, S, Hayashi, K, Kojima, C, Wong, H.L, Kawasaki, T, Shimamoto, K, Sato, M, Shimizu, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications
J.Biol.Chem., 285, 2010
|
|
2ZWH
 
 | | Model for the F-actin structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oda, T, Iwasa, M, Aihara, T, Maeda, Y, Narita, A. | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2017-10-11 | | Method: | FIBER DIFFRACTION (3.3 Å) | | Cite: | The nature of the globular- to fibrous-actin transition.
Nature, 457, 2009
|
|
3VPZ
 
 | |
5B2G
 
 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
1WYE
 
 | | Crystal structure of 2-keto-3-deoxygluconate kinase (form 1) from Sulfolobus Tokodaii | | Descriptor: | 2-keto-3-deoxygluconate kinase | | Authors: | Toyoda, T, Suzuki, K, Hossain, M.T, Koike, I, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2005-02-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus Tokodaii
To be Published
|
|
1IVI
 
 | | Crystal Structure of pig dihydrolipoamide dehydrogenase | | Descriptor: | dihydrolipoamide dehydrogenase | | Authors: | Toyoda, T, Kobayashi, R, Sekiguchi, T, Koike, K, Koike, M, Takenaka, A. | | Deposit date: | 2002-03-15 | | Release date: | 2003-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Crystallization and preliminary X-ray analysis of pig E3, lipoamide dehydrogenase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1JEH
 
 | | CRYSTAL STRUCTURE OF YEAST E3, LIPOAMIDE DEHYDROGENASE | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Toyoda, T, Suzuki, K, Sekigushi, T, Reed, J, Takenaka, A. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of eucaryotic E3, lipoamide dehydrogenase from yeast.
J.Biochem., 123, 1998
|
|
