1NLU
 
 | | Pseudomonas sedolisin (serine-carboxyl proteinase) complexed with two molecules of pseudo-iodotyrostatin | | Descriptor: | CALCIUM ION, PSEUDO-IODOTYROSTATIN, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Glodfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Two inhibitor molecules bound in the active site of Pseudomonas sedolisin: a model for the bi-product complex following cleavage of a peptide substrate.
Biochem.Biophys.Res.Commun., 314, 2004
|
|
6M9F
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Tyrostatin | | Descriptor: | CALCIUM ION, SEDOLISIN, SULFATE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M9C
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Pseudotyrostatin | | Descriptor: | ACETIC ACID, CALCIUM ION, Pseudotyrostatin, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M9D
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Chymostatin | | Descriptor: | CALCIUM ION, Chymostatin A, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8Y
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIPF | | Descriptor: | AIPF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8W
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIAF | | Descriptor: | AIAF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
1SN7
 
 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5PTI
 
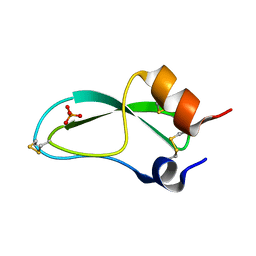 | |
9CBE
 
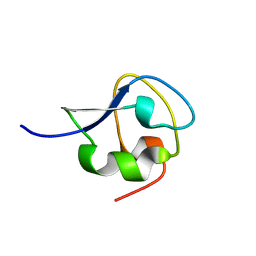 | |
5RSA
 
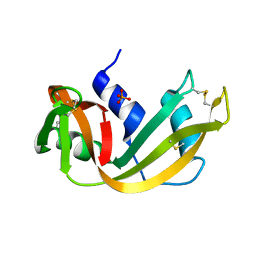 | |
1FMB
 
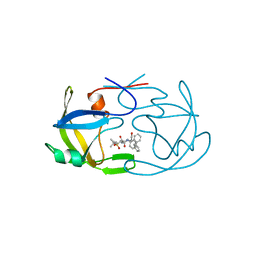 | | EIAV PROTEASE COMPLEXED WITH THE INHIBITOR HBY-793 | | Descriptor: | EIAV PROTEASE, [2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYL-VALINYL]-PHENYLALANINOL | | Authors: | Wlodawer, A, Gustchina, A, Zdanov, A, Kervinen, J. | | Deposit date: | 1996-02-27 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of equine infectious anemia virus proteinase complexed with an inhibitor.
Protein Sci., 5, 1996
|
|
1FIV
 
 | | STRUCTURE OF AN INHIBITOR COMPLEX OF PROTEINASE FROM FELINE IMMUNODEFICIENCY VIRUS | | Descriptor: | FIV PROTEASE, FIV PROTEASE INHIBITOR ACE-ALN-VAL-STA-GLU-ALN-NH2 | | Authors: | Wlodawer, A, Gustchina, A, Reshetnikova, L, Lubkowski, J, Zdanov, A. | | Deposit date: | 1995-05-04 | | Release date: | 1995-07-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus.
Nat.Struct.Biol., 2, 1995
|
|
6L4P
 
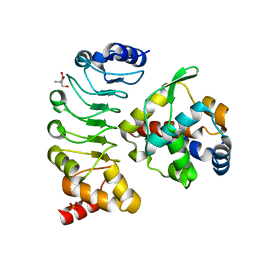 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
1GA6
 
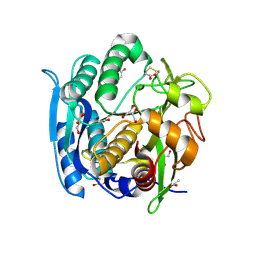 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF TYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | ACETATE ION, CALCIUM ION, FRAGMENT OF TYROSTATIN, ... | | Authors: | Wlodawer, A, Li, M, Dauter, Z, Gustchina, A, Uchida, K. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes.
Nat.Struct.Biol., 8, 2001
|
|
1GA4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH INHIBITOR PSEUDOIODOTYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | CALCIUM ION, GLYCEROL, PSEUDOIODOTYROSTATIN, ... | | Authors: | Wlodawer, A, Li, M, Dauter, Z, Gustchina, A, Uchida, K. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes.
Nat.Struct.Biol., 8, 2001
|
|
4XSH
 
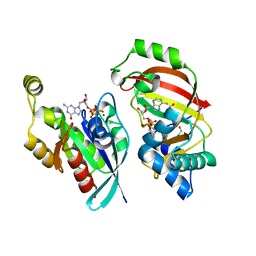 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4XSG
 
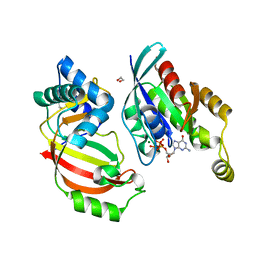 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-free state) | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
6RSA
 
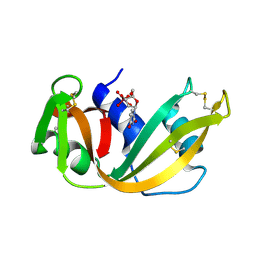 | |
7RSA
 
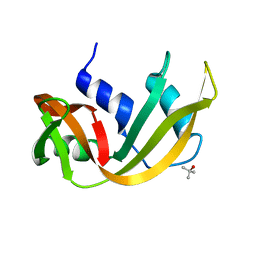 | |
6PTI
 
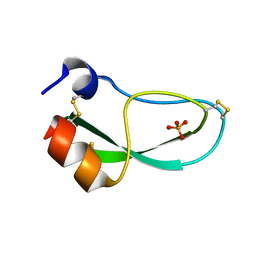 | |
5BWM
 
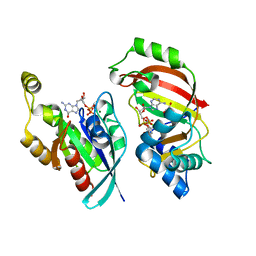 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-06-08 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
