1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
1EJ3
 
 | | CRYSTAL STRUCTURE OF AEQUORIN | | Descriptor: | AEQUORIN, C2-HYDROPEROXY-COELENTERAZINE | | Authors: | Head, J.F, Inouye, S, Teranishi, K, Shimomura, O. | | Deposit date: | 2000-02-29 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Nature, 405, 2000
|
|
5FLO
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(4-chloranylphenoxy)methyl]-1H-1,2,3,4-tetrazole, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5FLR
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5FRE
 
 | | Characterization of a novel CBM from Clostridium perfringens | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ribeiro, J, Pau, W, Pifferi, C, Renaudet, O, Varrot, A, Mahal, L.K, Imberty, A. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a High-Affinity Sialic Acid-Specific Cbm40 from Clostridium Perfringens and Engineering of a Divalent Form.
Biochem.J., 473, 2016
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2OJ1
 
 | | Disulfide-linked dimer of azurin N42C/M64E double mutant | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | de Jongh, T.E, Hoffmann, M, Einsle, O, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2007-01-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inter- and intramolecular electron transfer in modified azurin dimers
Eur.J.Inorg.Chem., 2007, 2007
|
|
5IC4
 
 | | Crystal structure of caspase-3 DEVE peptide complex | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
2NWL
 
 | | Crystal structure of GltPh in complex with L-Asp | | Descriptor: | ASPARTIC ACID, PALMITIC ACID, glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
5IFE
 
 | | Crystal structure of the human SF3b core complex | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Cretu, C, Dybkov, O, De Laurentiis, E, Will, C.L, Luhrmann, R, Pena, V. | | Deposit date: | 2016-02-25 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations.
Mol.Cell, 64, 2016
|
|
2OEB
 
 | | The crystal structure of gene product Af1862 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein | | Authors: | Zhang, R, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The crystal structure of gene product Af1862 from Archaeoglobus fulgidus
To be Published
|
|
2OFB
 
 | | Crystal structure of AVR4 (R112L/C122S)-BNA complex | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Avidin-related protein 4/5, FORMIC ACID | | Authors: | Livnah, O, Hayouka, R, Eisenberg-Domovich, Y. | | Deposit date: | 2007-01-03 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Critical importance of loop conformation to avidin-enhanced hydrolysis of an active biotin ester.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1D90
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH I.T MISMATCHED BASE PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*IP*GP*CP*TP*CP*C)-3') | | Authors: | Cruse, W.B.T, Aymani, J, Kennard, O, Brown, T, Jack, A.G.C, Leonard, G.A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs.
Nucleic Acids Res., 17, 1989
|
|
2ON3
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase | | Authors: | Dufe, V.T, Ingner, D, Heby, O, Khomutov, A.R, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
5HPZ
 
 | | type II water soluble Chl binding proteins | | Descriptor: | 13'2-hydroxyl-Chlorophyll a, Water-soluble chlorophyll protein | | Authors: | Bednarczyk, D, Dym, O, Prabahard, V, Noy, D. | | Deposit date: | 2016-01-21 | | Release date: | 2016-05-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Fine Tuning of Chlorophyll Spectra by Protein-Induced Ring Deformation.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1E31
 
 | | SURVIVIN DIMER H. SAPIENS | | Descriptor: | APOPTOSIS INHIBITOR SURVIVIN, COBALT (II) ION, ZINC ION | | Authors: | Chantalat, L, Skoufias, D.A, Margolis, R.L, Dideberg, O. | | Deposit date: | 2000-06-04 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of Human Survivin Reveals a Bow Tie-Shaped Dimer with Two Unusual Alpha-Helical Extensions
Mol.Cell, 6, 2000
|
|
5G17
 
 | | Bordetella Alcaligenes HDAH (T101A) bound to 9,9,9-trifluoro-8,8- dihydroxy-N-phenylnonanamide. | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
2O5V
 
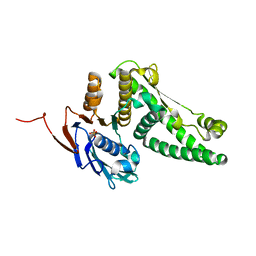 | | Recombination mediator RecF | | Descriptor: | DNA replication and repair protein recF, SULFATE ION | | Authors: | Korolev, S, Koroleva, O. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function.
Embo J., 26, 2007
|
|
5FNM
 
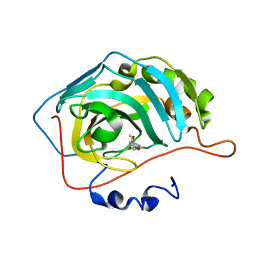 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | (~{E})-3-(4-methoxyphenyl)but-2-enoic acid, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5I6S
 
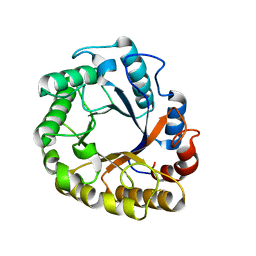 | | Crystal structure of an endoglucanase from Penicillium verruculosum | | Descriptor: | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, endoglucanase | | Authors: | Nemashklaov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinitsyn, A. | | Deposit date: | 2016-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum
To Be Published
|
|
2OF8
 
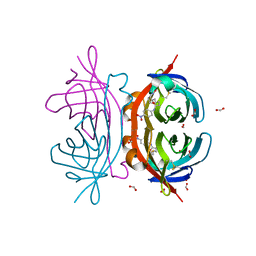 | | Crystal structure of AVR4 (D39A/C122S)-BNA complex | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Avidin-related protein 4/5, FORMIC ACID | | Authors: | Livnah, O, Hayouka, R, Eisenberg-Domovich, Y. | | Deposit date: | 2007-01-03 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Critical importance of loop conformation to avidin-enhanced hydrolysis of an active biotin ester.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2OCS
 
 | | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2) | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2, THIOCYANATE ION | | Authors: | Papagrigoriou, E, Phillips, C, Gileadi, C, Elkins, J, Salah, E, Berridge, G, Savitsky, P, Gorrec, F, Umeano, C, Ugochukwu, E, Gileadi, O, Burgess, N, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2)
To be Published
|
|
2OKQ
 
 | | Crystal structure of unknown conserved ybaA protein from Shigella flexneri | | Descriptor: | Hypothetical protein ybaA, SODIUM ION | | Authors: | Minasov, G, Vorontsov, I.I, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unknown conserved ybaA protein from Shigella flexneri
TO BE PUBLISHED
|
|
2OFA
 
 | | Crystal structure of apo AVR4 (R112L,C122S) | | Descriptor: | Avidin-related protein 4/5, FORMIC ACID | | Authors: | Livnah, O, Hayouka, R, Eisenberg-Domovich, Y. | | Deposit date: | 2007-01-03 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Critical importance of loop conformation to avidin-enhanced hydrolysis of an active biotin ester.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2O48
 
 | | Crystal structure of Mammalian Dimeric Dihydrodiol Dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, P-HYDROXYACETOPHENONE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2006-12-04 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis.
Proteins, 70, 2008
|
|
