8B0L
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | 分子名称: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | 分子名称: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | 分子名称: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | 分子名称: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | 著者 | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AN4
 
 | | MenT1 toxin (rv0078a) from Mycobacterium tuberculosis H37Rv | | 分子名称: | Bacterial toxin | | 著者 | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | 登録日 | 2022-08-04 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8AN5
 
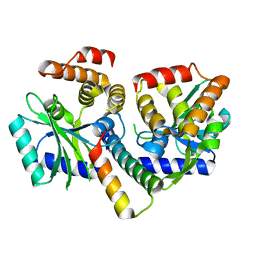 | | MenAT1 toxin-antitoxin complex (rv0078a-rv0078b) from Mycobacterium tuberculosis H37Rv | | 分子名称: | Bacterial toxin, Conserved protein | | 著者 | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | 登録日 | 2022-08-04 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8AFK
 
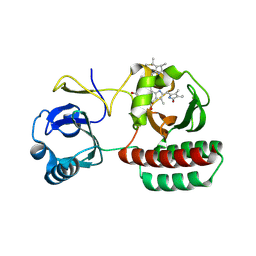 | | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin | | 分子名称: | Near-infrared fluorescent protein, PHYCOCYANOBILIN | | 著者 | Remeeva, A, Kovalev, K, Gushchin, I, Fonin, A, Turoverov, K, Stepanenko, O. | | 登録日 | 2022-07-18 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.015 Å) | | 主引用文献 | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin
To Be Published
|
|
8AH3
 
 | |
8AHX
 
 | |
8BHZ
 
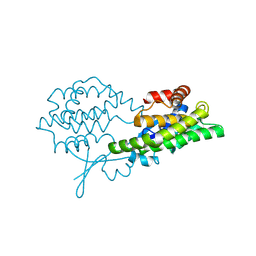 | |
7ZS5
 
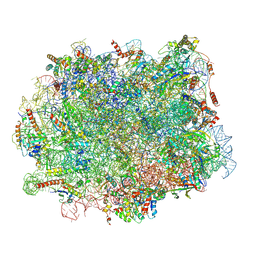 | | Structure of 60S ribosomal subunit from S. cerevisiae with eIF6 and tRNA | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-06 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZRS
 
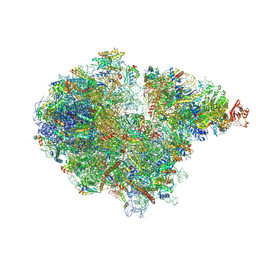 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUX
 
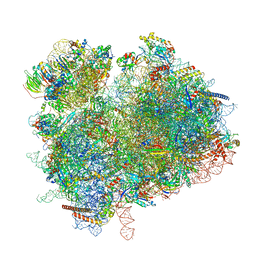 | | Collided ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUW
 
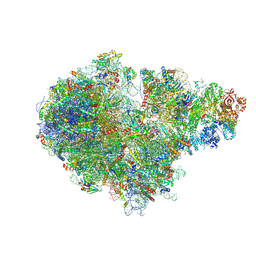 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZPQ
 
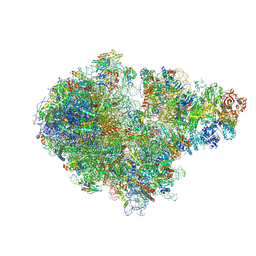 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-04-28 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZOK
 
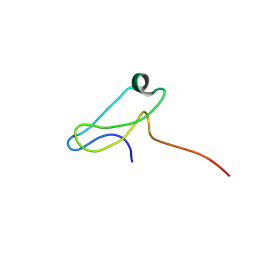 | | A novel molecular switch controls assembly of bacterial focal adhesions in response to changes in surface structure. | | 分子名称: | Adventurous gliding motility protein GltJ, ZINC ION | | 著者 | Attia, B, My, L, Castaing, J.P, Le Guenno, H, Espinosa, L, Schmidt, V, Nouailler, M, Bornet, O, Mignot, T, Elantak, L. | | 登録日 | 2022-04-25 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel molecular switch controls assembly of bacterial focal adhesions in response to changes in surface structure.
To Be Published
|
|
8BSS
 
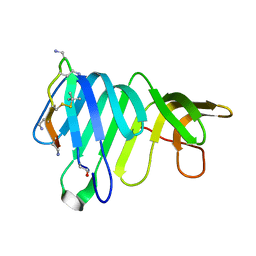 | | Solution Structure of thanatin-like derivative 5 in complex with E. coli LptA mutant Q62L | | 分子名称: | Lipopolysaccharide export system protein LptA, Thanatin-like derivative | | 著者 | Oi, K.K, Jurt, S, Moehle, K, Zerbe, O. | | 登録日 | 2022-11-26 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Peptidomimetic antibiotics disrupt the lipopolysaccharide transport bridge of drug-resistant Enterobacteriaceae.
Sci Adv, 9, 2023
|
|
8BQF
 
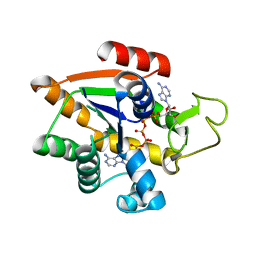 | | Adenylate Kinase L107I MUTANT | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | 著者 | Scheerer, D, Adkar, B.V, Bhattacharyya, S, Levy, D, Iljina, M, Iljina, I, Dym, O, Haran, G, Shakhnovich, E.I. | | 登録日 | 2022-11-21 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BOQ
 
 | |
8BQX
 
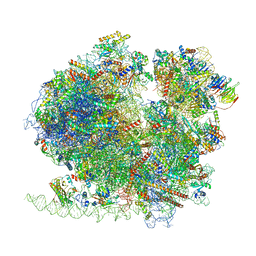 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BQD
 
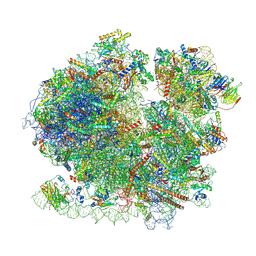 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BCX
 
 | |
7ZNQ
 
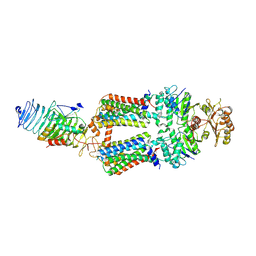 | | ABC transporter complex NosDFYL in GDN | | 分子名称: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | 著者 | Zhang, L, Mueller, C, Zipfel, S, Chami, M, Einsle, O. | | 登録日 | 2022-04-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
8AK3
 
 | |
8AK2
 
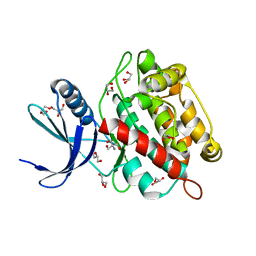 | | Drosophila melanogaster UNC89 Protein Kinase Domain 1 (apo) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Obscurin | | 著者 | Dorendorf, T, Zacharchenko, T, Mayans, O. | | 登録日 | 2022-07-29 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | PK1 from Drosophila obscurin is an inactive pseudokinase with scaffolding properties.
Open Biology, 13, 2023
|
|
