1J1G
 
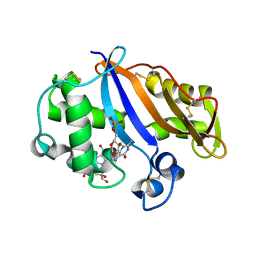 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
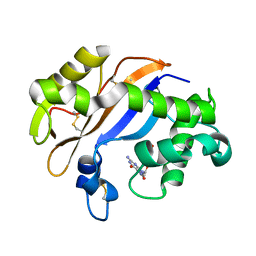 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
4IJ4
 
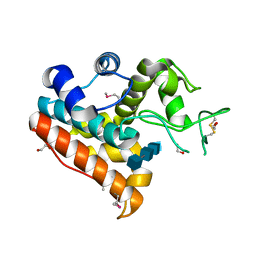 | | Crystal Structure of a Family GH19 chitinase from Bryum coronatum in complex with (GlcNAc)4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-12-21 | | Release date: | 2014-03-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
8YAN
 
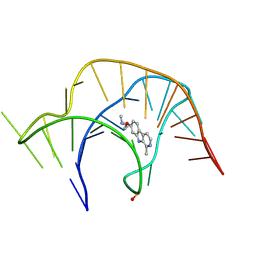 | |
8YAM
 
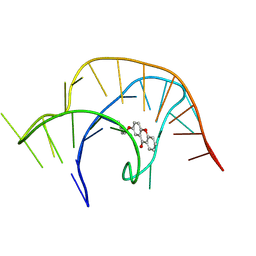 | |
4QTS
 
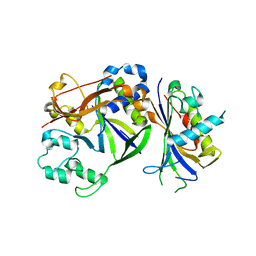 | |
4DWX
 
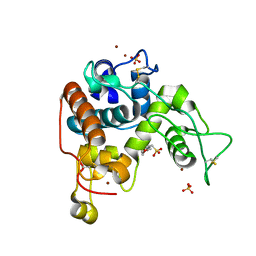 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Basic endochitinase C, SULFATE ION, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-27 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
4DYG
 
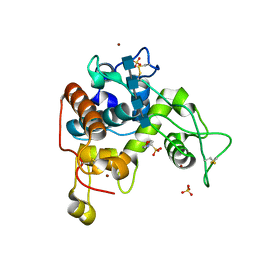 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds in complex with (GlcNAc)4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic endochitinase C, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
6AE2
 
 | |
6AE1
 
 | |
3WH1
 
 | | Crystal Structure of a Family GH19 Chitinase from Bryum coronatum in complex with (GlcNAc)4 at 1.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-08-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
3WIJ
 
 | | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Numata, T, Osawa, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3
To be Published
|
|
3W2W
 
 | | Crystal structure of the Cmr2dHD-Cmr3 subcomplex bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2012-12-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Cmr2-Cmr3 Subcomplex in the CRISPR-Cas RNA Silencing Effector Complex.
J.Mol.Biol., 425, 2013
|
|
3W2V
 
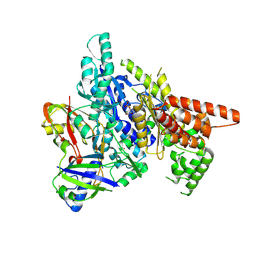 | |
2D1P
 
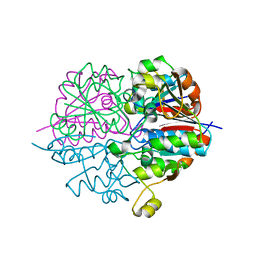 | | crystal structure of heterohexameric TusBCD proteins, which are crucial for the tRNA modification | | Descriptor: | Hypothetical UPF0116 protein yheM, Hypothetical UPF0163 protein yheN, Hypothetical protein yheL, ... | | Authors: | Numata, T, Fukai, S, Ikeuchi, Y, Suzuki, T, Nureki, O. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Sulfur Relay to RNA Mediated by Heterohexameric TusBCD Complex
Structure, 14, 2006
|
|
2DER
 
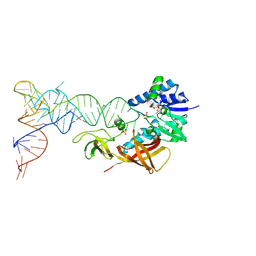 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DET
 
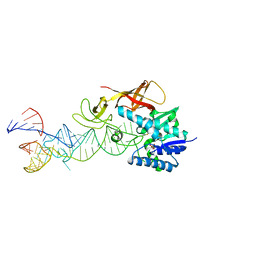 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
7E9I
 
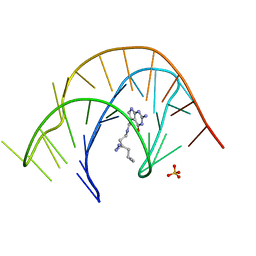 | | Crystal structure of a class I PreQ1 riboswitch aptamer (wild-type) complexed with a cognate ligand-derived photoaffinity probe | | Descriptor: | 2-azanyl-5-[[2-(3-but-3-ynyl-1,2-diazirin-3-yl)ethylamino]methyl]-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one, 33-mer RNA, SULFATE ION | | Authors: | Numata, T, Schneekloth, J.S. | | Deposit date: | 2021-03-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites.
Nat Commun, 12, 2021
|
|
7E9E
 
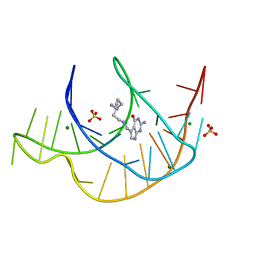 | | Crystal structure of a class I PreQ1 riboswitch aptamer (ab13-14) complexed with a cognate ligand-derived photoaffinity probe | | Descriptor: | 2-azanyl-5-[[2-(3-but-3-ynyl-1,2-diazirin-3-yl)ethylamino]methyl]-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one, 33-mer RNA, MAGNESIUM ION, ... | | Authors: | Numata, T, Schneekloth, J.S. | | Deposit date: | 2021-03-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites.
Nat Commun, 12, 2021
|
|
2DEU
 
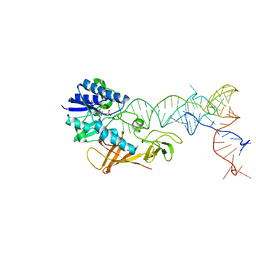 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the adenylated intermediate state | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
6E1V
 
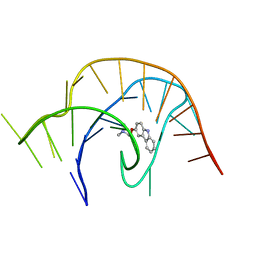 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 3: 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1S
 
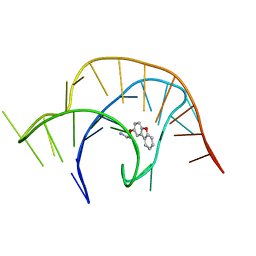 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1T
 
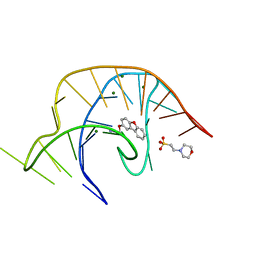 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1W
 
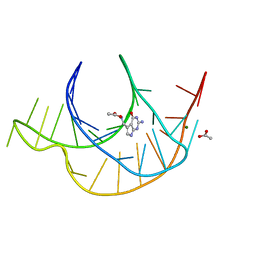 | | Crystal structure of a class I PreQ1 riboswitch complexed with PreQ1 | | Descriptor: | 2-amino-5-(aminomethyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1U
 
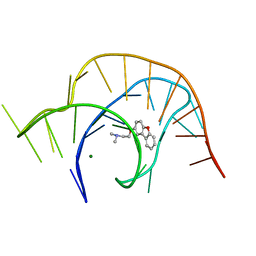 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 2: 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine, MAGNESIUM ION, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
