5U3Y
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 9 | | Descriptor: | 6-[2-({cyclopropyl[4-(furan-2-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U3W
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 7 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(cyclopropyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U45
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 14 | | Descriptor: | 6-(2-{[cyclopropyl(3'-fluoro[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8SMS
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase, FabB, and cerulenin crosslinker-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-{2-[(2R)-2-hydroxy-4-oxododecanamido]ethyl}-beta-alaninamide | | Authors: | Jiang, Z, Chen, A, Chen, J, Sekhon, A, Louie, G.V, Noel, J.P, La Clair, J.J, Burkart, M.D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Masked cerulenin enables a dual-site selective protein crosslink.
Chem Sci, 14, 2023
|
|
7SQI
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SZ9
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6VKZ
 
 | |
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4JYP
 
 | | crystal Structure of KAI2 Apo form | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYM
 
 | | crystal Structure of KAI2 in complex with 3-methyl-2H-furo[2,3-c]pyran-2-one | | Descriptor: | 3-methyl-2H-furo[2,3-c]pyran-2-one, Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-30 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6VL0
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | Descriptor: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3C1O
 
 | |
3C3X
 
 | |
5BUK
 
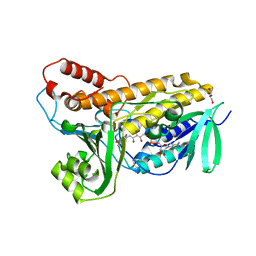 | | Structure of flavin-dependent chlorinase Mpy16 | | Descriptor: | FADH2-dependent halogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BUL
 
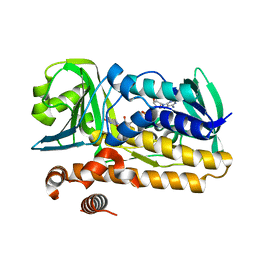 | | Structure of flavin-dependent brominase Bmp2 triple mutant Y302S F306V A345W | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase triple mutant | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9784 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BVA
 
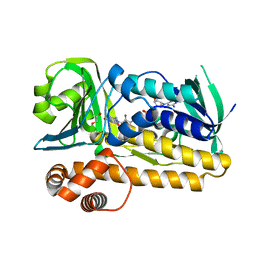 | | Structure of flavin-dependent brominase Bmp2 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TAD
 
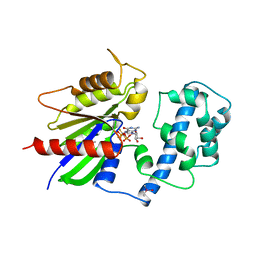 | | GTPASE MECHANISM OF GPROTEINS FROM THE 1.7-ANGSTROM CRYSTAL STRUCTURE OF TRANSDUCIN ALPHA-GDP-ALF4- | | Descriptor: | CACODYLATE ION, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sondek, J, Lambright, D.G, Noel, J.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1995-01-05 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4.
Nature, 372, 1994
|
|
5EAU
 
 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION, TRIFLUOROFURNESYL DIPHOSPHATE | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
5EAT
 
 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM WITH SUBSTRATE ANALOG FARNESYL HYDROXYPHOSPHONATE | | Descriptor: | 1-HYDROXY-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENE PHOSPHONIC ACID, 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-07-24 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
5EAS
 
 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-06-19 | | Release date: | 1997-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
3B5I
 
 | | Crystal structure of Indole-3-acetic Acid Methyltransferase | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase-like protein | | Authors: | Ferrer, J.-L, Noel, J.P. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural, Biochemical, and Phylogenetic Analyses Suggest That Indole-3-Acetic Acid Methyltransferase Is an Evolutionarily Ancient Member of the SABATH Family.
Plant Physiol., 146, 2008
|
|
6CJN
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95T mutation | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CIG
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
6CJO
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
3K52
 
 | |
