5JH9
 
 | | Crystal structure of prApe1 | | Descriptor: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGF
 
 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
7W3O
 
 | | Crystal structure of human CYB5R3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3 soluble form | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
7W3N
 
 | | Crystal structure of Ufm1 fused to UFBP1 UFIM | | Descriptor: | UFBP1 peptide,Ubiquitin-fold modifier 1 | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
7VEC
 
 | |
2ZPN
 
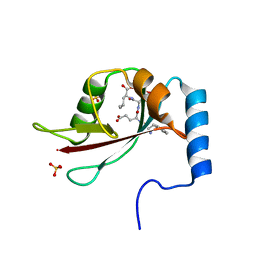 | |
3VH3
 
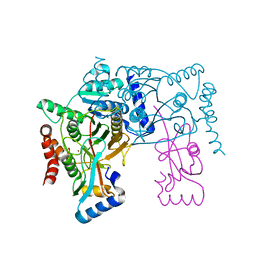 | | Crystal structure of Atg7CTD-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VP7
 
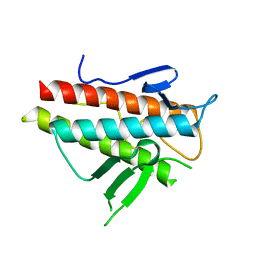 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
3W1S
 
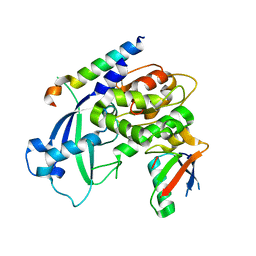 | | Crystal structure of Saccharomyces cerevisiae Atg12-Atg5 conjugate bound to the N-terminal domain of Atg16 | | Descriptor: | Autophagy protein 16, Autophagy protein 5, Ubiquitin-like protein ATG12 | | Authors: | Noda, N.N, Fujioka, Y, Hanada, T, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Embo Rep., 14, 2013
|
|
3VH2
 
 | |
3VH1
 
 | |
3VXW
 
 | |
3VH4
 
 | | Crystal structure of Atg7CTD-Atg8-MgATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
5H9W
 
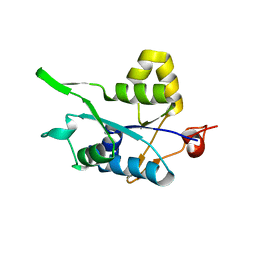 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
5H9V
 
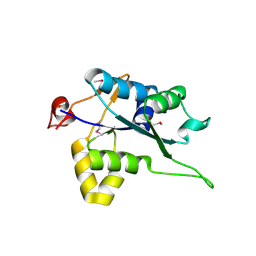 | | Crystal structure of Regnase PIN domain, form I | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
4P1W
 
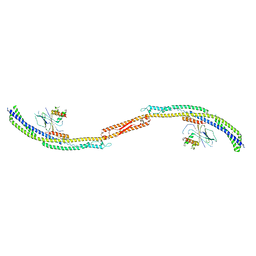 | |
4P1N
 
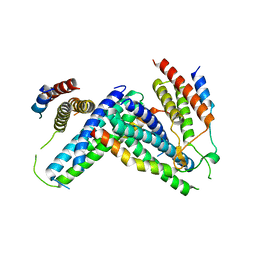 | | Crystal structure of Atg1-Atg13 complex | | Descriptor: | Atg1 tMIT, Atg13 MIM | | Authors: | Fujioka, Y, Noda, N.N. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of starvation-induced assembly of the autophagy initiation complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5JGE
 
 | |
5JHC
 
 | |
5JHF
 
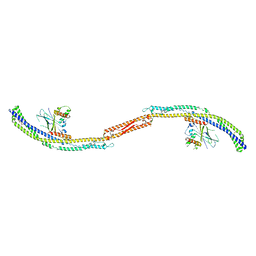 | |
7BRN
 
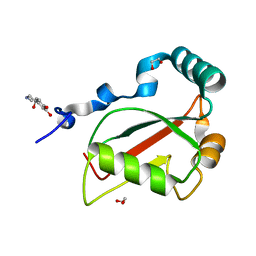 | | Crystal structure of Atg40 AIM fused to Atg8 | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 40,Autophagy-related protein 8, L-EPINEPHRINE | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRT
 
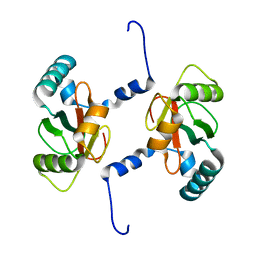 | | Crystal structure of Sec62 LIR fused to GABARAP | | Descriptor: | Translocation protein SEC62,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRQ
 
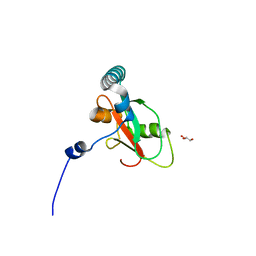 | | Crystal structure of human FAM134B LIR fused to human GABARAP | | Descriptor: | GLYCEROL, Reticulophagy regulator 1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRU
 
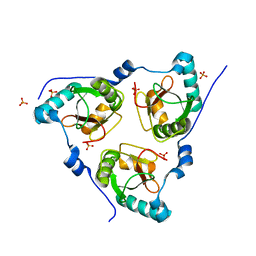 | | Crystal structure of human RTN3 LIR fused to human GABARAP | | Descriptor: | PHOSPHATE ION, Reticulon-3,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7YDO
 
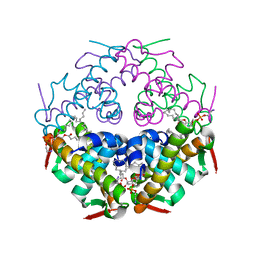 | | Crystal structure of Atg44 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Uncharacterized protein C26A3.14c | | Authors: | Maruyama, T, Noda, N.N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy.
Mol.Cell, 83, 2023
|
|
