3KLJ
 
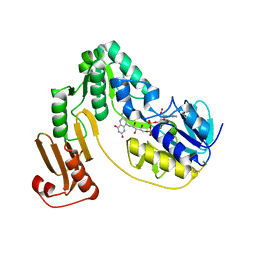 | | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) | | Authors: | Nishikawa, K, Shomura, Y, Kawasaki, S, Niimura, Y, Higuchi, Y. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum: a key component of the dioxygen scavenging system in obligatory anaerobes.
Proteins, 78, 2010
|
|
7VXQ
 
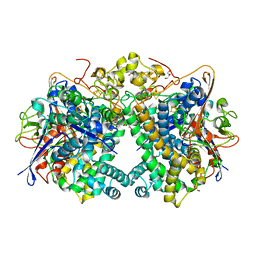 | | The Carbon Monoxide Complex of [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 | | Descriptor: | CARBON MONOXIDE, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Higuchi, K, Imanishi, T, Higuchi, Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and spectroscopic characterization of CO inhibition of [NiFe]-hydrogenase from Citrobacter sp. S-77.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4HPC
 
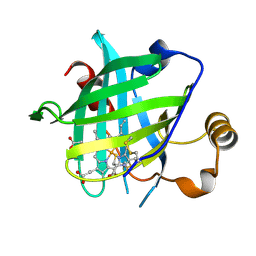 | |
4HPB
 
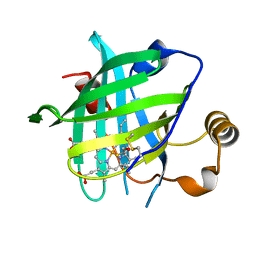 | |
4HPD
 
 | |
4HPA
 
 | |
5XVB
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVC
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
5XLG
 
 | | Crystal structure of anaerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLE
 
 | | Crystal structure of anaerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLF
 
 | | Crystal structure of aerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
2D57
 
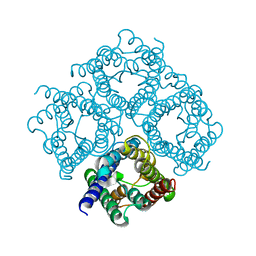 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
8IHI
 
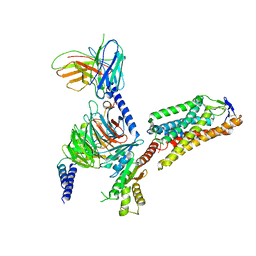 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
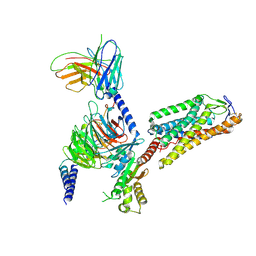 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
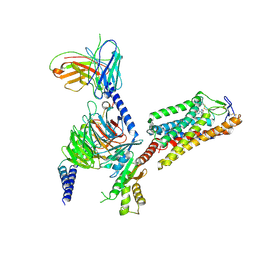 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
4U9I
 
 | | High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
