4BCY
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, mutation H43F | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Awad, W, Saraboji, K, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5NVP
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5ODL
 
 | | Single-stranded DNA-binding protein from bacteriophage Enc34 in complex with ssDNA | | Descriptor: | GLYCEROL, SODIUM ION, oligo(T), ... | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2017-07-05 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34.
Sci Rep, 7, 2017
|
|
5ODJ
 
 | | Single-stranded DNA-binding protein from bacteriophage Enc34 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Single-stranded DNA-binding protein | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2017-07-05 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34.
Sci Rep, 7, 2017
|
|
5ODK
 
 | | Single-stranded DNA-binding protein from bacteriophage Enc34, C-terminal truncation | | Descriptor: | GLYCEROL, PHOSPHATE ION, single-stranded DNA-binding protein | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2017-07-05 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34.
Sci Rep, 7, 2017
|
|
2YGV
 
 | | Conserved N-terminal domain of the yeast Histone Chaperone Asf1 in complex with the C-terminal fragment of Rad53 | | Descriptor: | GLYCEROL, HISTONE CHAPERONE ASF1, SERINE/THREONINE-PROTEIN KINASE RAD53, ... | | Authors: | Jiao, Y, Seeger, K, Murciano, B, Ledu, M.H, Charbonnier, J.B, Legrand, P, Lautrette, A, Gaubert, A, Mousson, F, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Surprising Complexity of the Asf1 Histone Chaperone-Rad53 Kinase Interaction
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5NWW
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
4A3V
 
 | | yeast regulatory particle proteasome assembly chaperone Hsm3 in complex with Rpt1 C-terminal fragment | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 7 HOMOLOG, DNA MISMATCH REPAIR PROTEIN HSM3, LINKER | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A3T
 
 | | yeast regulatory particle proteasome assembly chaperone Hsm3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN HSM3 | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HV0
 
 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
2IPI
 
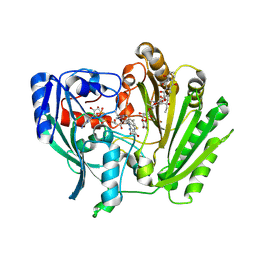 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4KWI
 
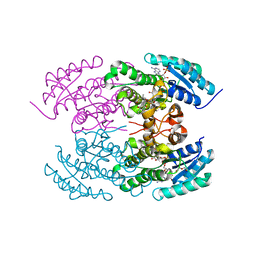 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP and 11-deoxy-6-oxylandomycinone | | Descriptor: | 1,8-dihydroxy-3-methyltetraphene-6,7,12(5H)-trione, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
4BCZ
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form. | | Descriptor: | SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Saraboji, K, Awad, W, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1RZS
 
 | |
1R1K
 
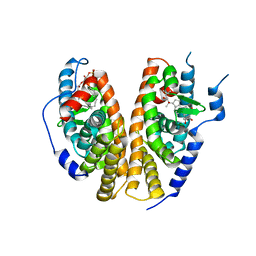 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
4EX6
 
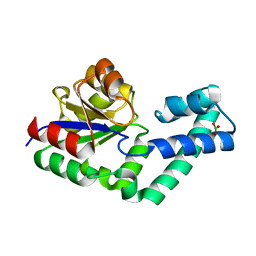 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX7
 
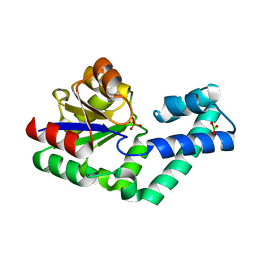 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
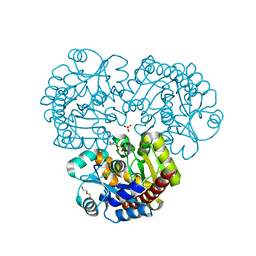 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX9
 
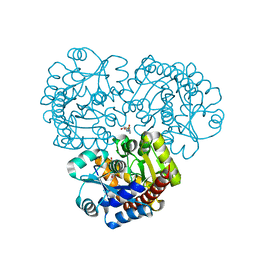 | | Crystal structure of the prealnumycin C-glycosynthase AlnA in complex with ribulose 5-phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AlnA, CALCIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2UYL
 
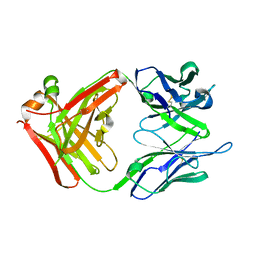 | | Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 | | Descriptor: | MONOCLONAL ANTIBODY F-22-30 | | Authors: | Ahmed, F, Haouz, A, Nato, F, Fournier, J.M, Alzari, P.M. | | Deposit date: | 2007-04-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Proteins, 70, 2008
|
|
1R20
 
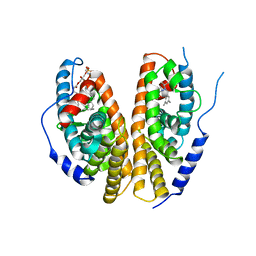 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI06830 | | Descriptor: | ECDYSONE RECEPTOR, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, N-(TERT-BUTYL)-3,5-DIMETHYL-N'-[(5-METHYL-2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)CARBONYL]BENZOHYDRAZIDE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-25 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
