1TQ8
 
 | | Crystal Structure of protein Rv1636 from Mycobacterium tuberculosis H37Rv | | Descriptor: | hypothetical protein Rv1636 | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hypothetical protein Rv1636 from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2L57
 
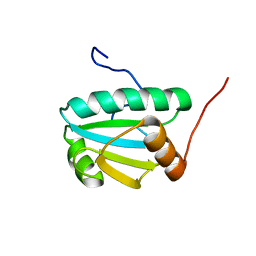 | | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens
To be Published
|
|
1TVF
 
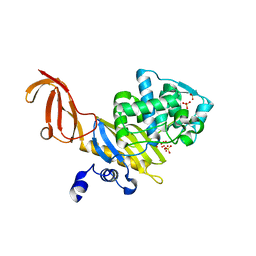 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | Descriptor: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
1U05
 
 | |
1U6M
 
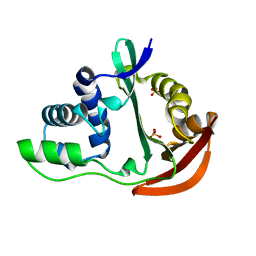 | | The crystal structure of acetyltransferase | | Descriptor: | SULFATE ION, acetyltransferase, GNAT family | | Authors: | Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of acetyltransferase, GNAT family from Enterococcus faecalis
To be Published
|
|
1VQV
 
 | |
1TT7
 
 | |
1TLT
 
 | | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG) | | Descriptor: | PUTATIVE OXIDOREDUCTASE (VIRULENCE FACTOR mviM HOMOLOG), SULFATE ION | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG)
To be Published
|
|
1T5J
 
 | |
1XD7
 
 | |
2HCM
 
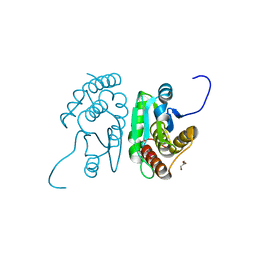 | | Crystal structure of mouse putative dual specificity phosphatase complexed with zinc tungstate, New York Structural Genomics Consortium | | Descriptor: | Dual specificity protein phosphatase, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1TZZ
 
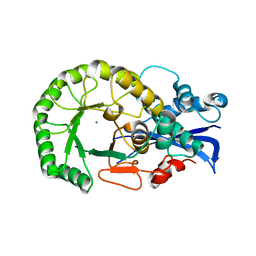 | | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum | | Descriptor: | Hypothetical protein L1841, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum
To be Published
|
|
1JG8
 
 | | Crystal Structure of Threonine Aldolase (Low-specificity) | | Descriptor: | CALCIUM ION, L-allo-threonine aldolase, SODIUM ION | | Authors: | Kielkopf, C.L, Bonanno, J, Ray, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-23 | | Release date: | 2001-07-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Low-specificity Threonine Aldolase, a Key Enzyme in Glycine Biosynthesis
To be published
|
|
1JZT
 
 | |
1TXN
 
 | |
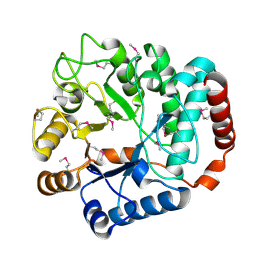
1TR9
 
 | |
1WVI
 
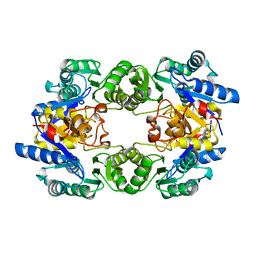 | |
1HQZ
 
 | | Cofilin homology domain of a yeast actin-binding protein ABP1P | | Descriptor: | ACTIN-BINDING PROTEIN | | Authors: | Strokopytov, B.V, Fedorov, A.A, Mahoney, N, Drubin, D.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-12-20 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2L5L
 
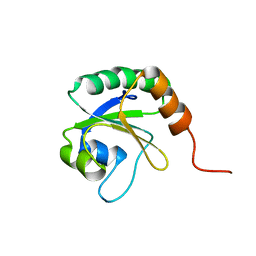 | | Solution Structure of Thioredoxin from Bacteroides Vulgatus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Thioredoxin from Bacteroides Vulgatus
To be Published
|
|
2L5O
 
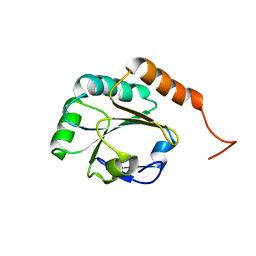 | | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis | | Descriptor: | Putative thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis
To be Published
|
|
1RXD
 
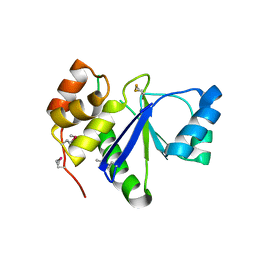 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1X94
 
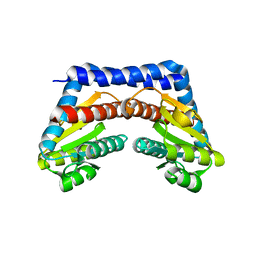 | |
1TSJ
 
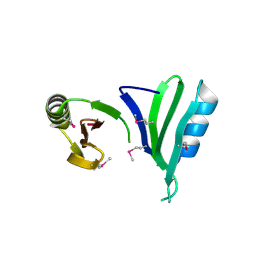 | |
1XEA
 
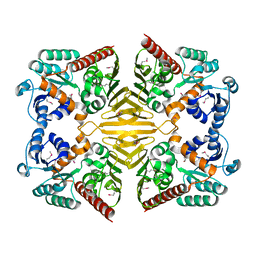 | | Crystal structure of a Gfo/Idh/MocA family oxidoreductase from Vibrio cholerae | | Descriptor: | NICKEL (II) ION, Oxidoreductase, Gfo/Idh/MocA family | | Authors: | R Rajashankar, K, Reynes, J.A, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-09-09 | | Release date: | 2004-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a Gfo/Idh/MocA family oxidoreductase from Vibrio cholerae
To be Published
|
|
1JFI
 
 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
