2QYZ
 
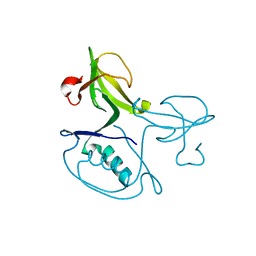 | | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88 | | Descriptor: | Uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Wasserman, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-15 | | Release date: | 2007-08-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88.
To be Published
|
|
3H14
 
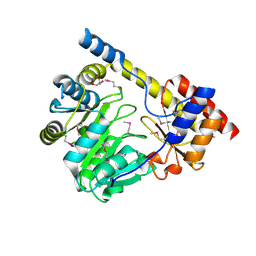 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3H9M
 
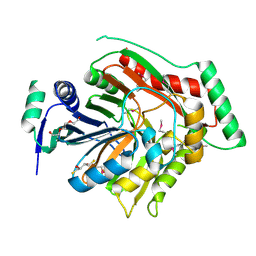 | | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, TRIETHYLENE GLYCOL, p-aminobenzoate synthetase, ... | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Do, J, Bain, K, Rutter, M, Gheyi, T, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii
To be Published
|
|
3LTE
 
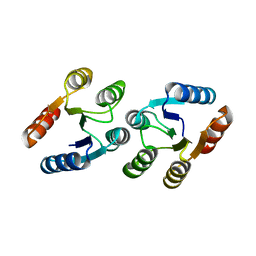 | | CRYSTAL STRUCTURE OF RESPONSE REGULATOR (SIGNAL RECEIVER DOMAIN) FROM Bermanella marisrubri | | Descriptor: | GLYCEROL, PHOSPHATE ION, Response regulator | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RESPONSE REGULATOR SIGNAL RECEIVER DOMAIN FROM Bermanella marisrubri RED65
To be Published
|
|
3HG7
 
 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3DTD
 
 | | Crystal structure of invasion associated protein b from bartonella henselae | | Descriptor: | GLYCEROL, Invasion-associated protein B | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Slocombe, A, Groshong, C, Koss, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Invasion Associated Protein B from Bartonella Henselae.
To be Published
|
|
3MOG
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
2NR4
 
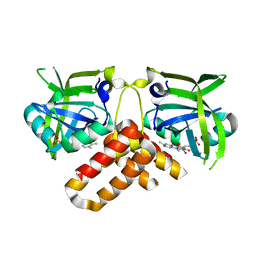 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
2OVL
 
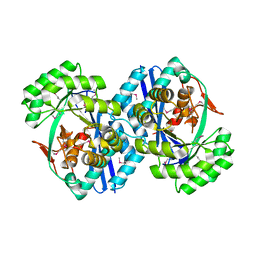 | |
2OY9
 
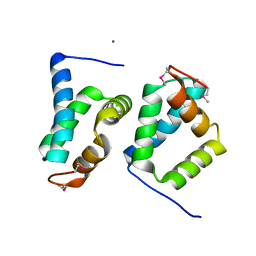 | |
2OO6
 
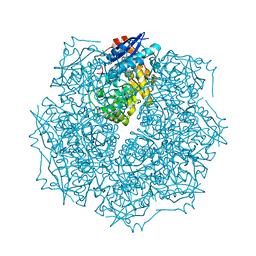 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 | | Descriptor: | Putative L-alanine-DL-glutamate epimerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Sridhar, V, Freeman, J, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400
To be Published
|
|
2OQH
 
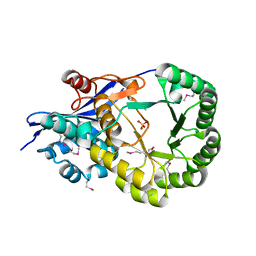 | |
2NLY
 
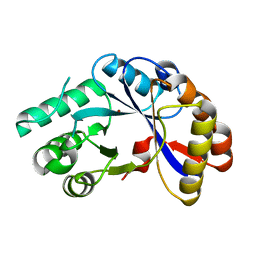 | | Crystal structure of protein BH1492 from Bacillus halodurans, Pfam DUF610 | | Descriptor: | Divergent polysaccharide deacetylase hypothetical protein, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein BH1492 from Bacillus halodurans C-125
To be Published
|
|
2P67
 
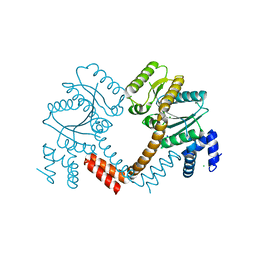 | | Crystal structure of LAO/AO transport system kinase | | Descriptor: | CHLORIDE ION, LAO/AO transport system kinase, SODIUM ION | | Authors: | Ramagopal, U.A, Adams, J, Rodgers, L, Toro, R, Bain, K, Rutter, M, Schwinn, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LAO/AO transport system kinase
To be Published
|
|
2PTF
 
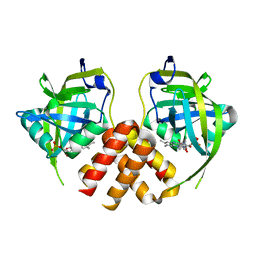 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
3K4W
 
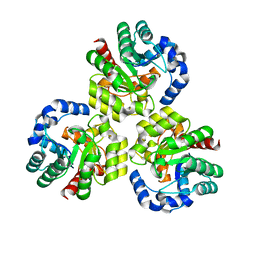 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
2PLG
 
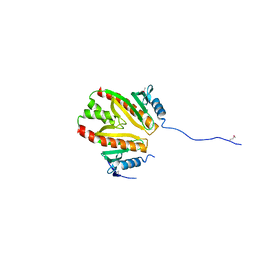 | |
2Q6E
 
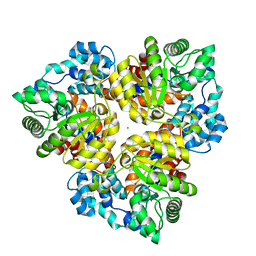 | | Crystal structure of glucuronate isomerase from Bacillus halodurans complexed with Zn | | Descriptor: | BH0493 protein, CHLORIDE ION, SODIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Rauschel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of glucuronate isomerase from Bacillus halodurans complexed with Zn.
To be Published
|
|
2PWW
 
 | | Crystal structure of ABC2387 from Bacillus clausii | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein | | Authors: | Ramagopal, U.A, Freeman, J, Lau, C, Toro, R, Bain, K, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of ABC2387 from Bacillus clausii.
To be Published
|
|
2POZ
 
 | |
3GZ4
 
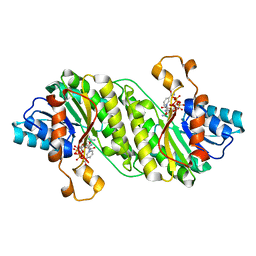 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3H12
 
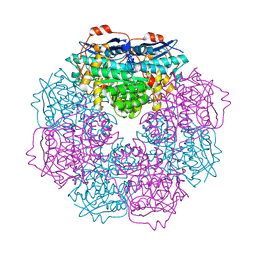 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
2PW9
 
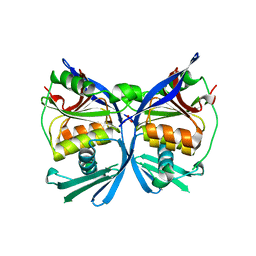 | | Crystal structure of a putative formate dehydrogenase accessory protein from Desulfotalea psychrophila | | Descriptor: | Putative formate dehydrogenase accessory protein, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Logan, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative formate dehydrogenase accessory protein from Desulfotalea psychrophila.
To be Published
|
|
2PB9
 
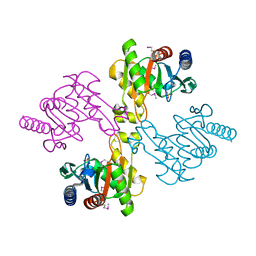 | |
