5U90
 
 | | Crystal structure of Co-CAO1 in complex with resveratrol | | Descriptor: | COBALT (II) ION, Carotenoid oxygenase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sui, X, Palczewski, k, Banerjee, S, Kiser, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center.
Biochemistry, 56, 2017
|
|
5U8X
 
 | | Crystal structure of Fe-CAO1 | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Carotenoid oxygenase 1, ... | | Authors: | Sui, X, Palczewski, K, Banerjee, S, Kiser, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center.
Biochemistry, 56, 2017
|
|
1ZCH
 
 | | Structure of the hypothetical oxidoreductase YcnD from Bacillus subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morokutti, A, Lyskowski, A, Sollner, S, Pointner, E, Fitzpatrick, T.B, Kratky, C, Gruber, K, Macheroux, P. | | Deposit date: | 2005-04-12 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Function of YcnD from Bacillus subtilis, a Flavin-Containing Oxidoreductase(,).
Biochemistry, 44, 2005
|
|
5U97
 
 | | Crystal structure of Co-CAO1 in complex with piceatannol | | Descriptor: | BENZOIC ACID, COBALT (II) ION, Carotenoid oxygenase 1, ... | | Authors: | Sui, X, Palczewski, K, Banerjee, S, Kiser, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center.
Biochemistry, 56, 2017
|
|
2OHY
 
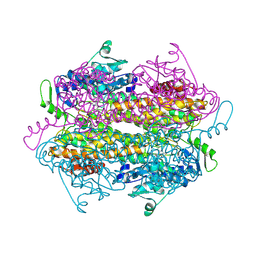 | |
1U8E
 
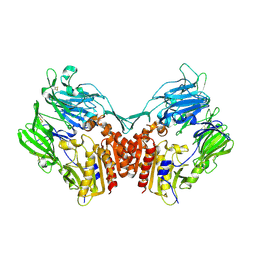 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
6QZJ
 
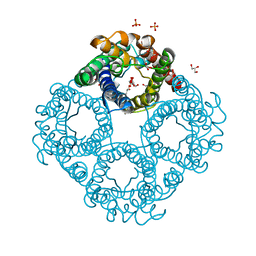 | | Crystal structure of human Aquaporin 7 at 2.2 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
4AQH
 
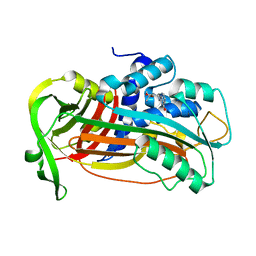 | | Plasminogen activator inhibitor type-1 in complex with the inhibitor AZ3976 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR 1, TERT-BUTYL 3-[(4-OXO-3H-PYRIDO[2,3-D]PYRIMIDIN-2-YL)AMINO]AZETIDINE-1-CARBOXYLATE | | Authors: | Fjellstrom, O, Deinum, J, Sjogren, T, Johansson, C, Geschwindner, S, Nerme, V, Legnehed, A, McPheat, J, Olsson, K, Bodin, C, Gustafsson, D. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a Small Molecule Inhibitor of Plasminogen Activator Inhibitor Type 1 that Accelerates the Transition Into the Latent Conformation
J.Biol.Chem., 288, 2013
|
|
2W0R
 
 | | Crystal structure of the mutated N263D YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
5A3L
 
 | | Structure of Cea1A in complex with N-Acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
6QZI
 
 | | Crystal structure of human Aquaporin 7 at 1.9 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
1POZ
 
 | | SOLUTION STRUCTURE OF THE HYALURONAN BINDING DOMAIN OF HUMAN CD44 | | Descriptor: | CD44 antigen | | Authors: | Teriete, P, Banerji, S, Blundell, C.D, Kahmann, J.D, Pickford, A.R, Wright, A.J, Campbell, I.D, Jackson, D.G, Day, A.J. | | Deposit date: | 2003-06-16 | | Release date: | 2004-03-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44.
Mol.Cell, 13, 2004
|
|
5A3M
 
 | | Structure of Cea1A in complex with Chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CEA1, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
1UUH
 
 | | Hyaluronan binding domain of human CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Teriete, P, Banerji, S, Noble, M, Blundell, C, Wright, A, Pickford, A, Lowe, E, Mahoney, D, Tammi, M, Kahmann, J, Campbell, I, Day, A, Jackson, D. | | Deposit date: | 2003-12-19 | | Release date: | 2004-03-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor Cd44
Mol.Cell, 13, 2004
|
|
1T3Q
 
 | | Crystal structure of quinoline 2-Oxidoreductase from Pseudomonas Putida 86 | | Descriptor: | DIOXOSULFIDOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonin, I, Martins, B.M, Purvanov, V, Fetzner, S, Huber, R, Dobbek, H. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
STRUCTURE, 12, 2004
|
|
4G95
 
 | | hDHFR-OAG binary complex | | Descriptor: | 6-{[(2,5-dichlorophenyl)amino]methyl}pyrido[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Pace, J, Queener, S, Adair, O, Gangjee, A. | | Deposit date: | 2012-07-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Kinetic and structural analysis for potent antifolate inhibition of Pneumocystis jirovecii, Pneumocystis carinii, and human dihydrofolate reductases and their active-site variants.
Antimicrob.Agents Chemother., 57, 2013
|
|
4OTJ
 
 | | The complex of murine cyclooxygenase-2 with a conjugate of indomefathin and podophyllotoxin, N-{(succinylpodophyllotoxinyl)but-4-yl}-2-{1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl}acetamide | | Descriptor: | (5S,5aS,8aS,9S)-8-oxo-9-(3,4,5-trimethoxyphenyl)-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4-{[4-({[1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl]acetyl}amino)butyl]amino}-4-oxobutanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Antitumor Activity of Cytotoxic Cyclooxygenase-2 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
2XJR
 
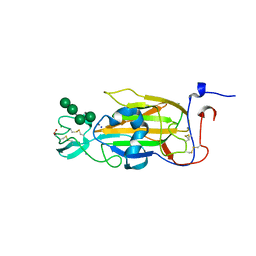 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D2-D3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4OTY
 
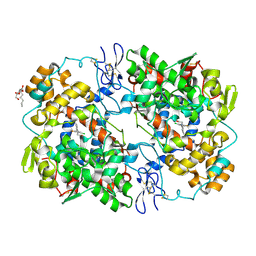 | | Crystal structure of lumiracoxib bound to the apo-mouse-cyclooxygenase-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Windsor, M.A, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Exploring the molecular determinants of substrate-selective inhibition of cyclooxygenase-2 by lumiracoxib.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3CD2
 
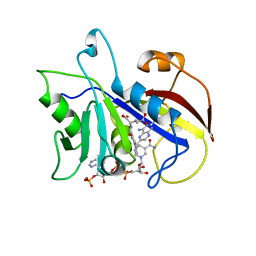 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
2XJT
 
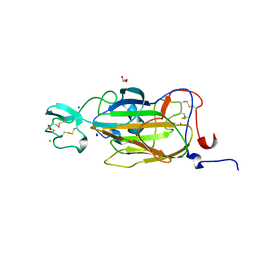 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJP
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WRZ
 
 | | Crystal structure of an arabinose binding protein with designed serotonin binding site in open, ligand-free state | | Descriptor: | L-ARABINOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Schreier, B, Stumpp, C, Wiesner, S, Hocker, B. | | Deposit date: | 2009-09-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Computational Design of Ligand Binding is not a Solved Problem
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XJQ
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GLYCEROL, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
