5EJK
 
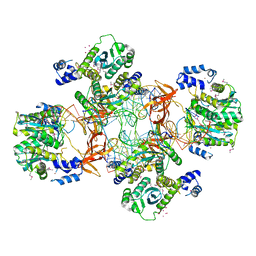 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
6FX4
 
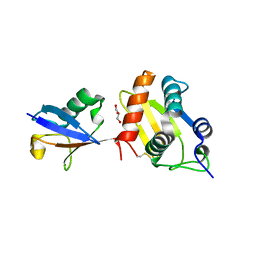 | |
6FU6
 
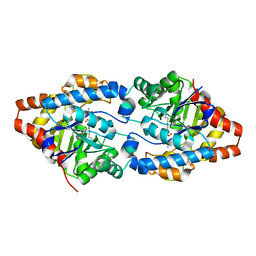 | | Phosphotriesterase PTE_C23_2 | | Descriptor: | FORMIC ACID, POLYACRYLIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
6G3M
 
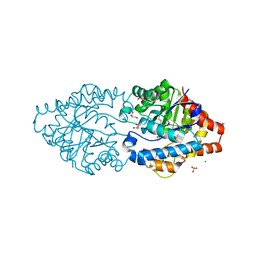 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
6G1J
 
 | | Phosphotriesterase PTE_C23M_1 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphotriesterase
PTE_C23M_1
To Be Published
|
|
6FWE
 
 | | Phosphotriesterase PTE_C23_6 | | Descriptor: | 1-[methoxy(methyl)phosphoryl]oxyethane, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Phosphotriesterase
PTE_C23_6
To Be Published
|
|
6FRZ
 
 | | Phosphotriesterase PTE_A53_7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methylcyclohexane-1,1,3,3-tetrol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase
PTE_A53_7
To Be Published
|
|
6WLG
 
 | | Ints3 C-terminal Domain | | Descriptor: | Integrator complex subunit 3 | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Structural basis for multifunctional roles of human Ints3 C-terminal domain.
J.Biol.Chem., 296, 2020
|
|
5E33
 
 | | Structure of human DPP3 in complex with met-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, Met-enkephalin, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5W58
 
 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | Descriptor: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
5E3C
 
 | | Structure of human DPP3 in complex with hemorphin like opioid peptide IVYPW | | Descriptor: | Dipeptidyl peptidase 3, IVYPW, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5EKP
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5F15
 
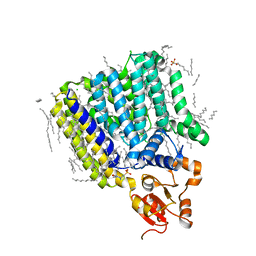 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | Descriptor: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
5EZM
 
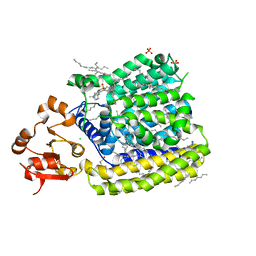 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
6E5T
 
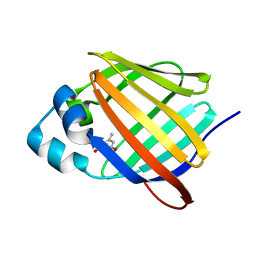 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
5FLJ
 
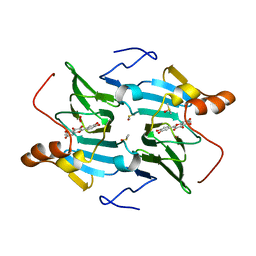 | | enzyme-substrate-dioxygen complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, NICKEL (II) ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5FLI
 
 | | enzyme-substrate complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NICKEL (II) ION, QUERCETINASE QUED | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-On O2-Ni Complex.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5EHH
 
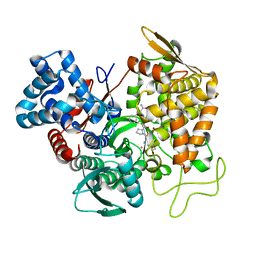 | | Structure of human DPP3 in complex with endomorphin-2. | | Descriptor: | Dipeptidyl peptidase 3, Endomorphin-2, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EGY
 
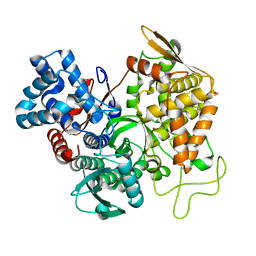 | | Structure of ligand free human DPP3 in closed form. | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, ZINC ION | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.741 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5DWZ
 
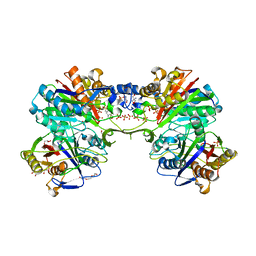 | | Structural and functional characterization of PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-(Acyl carrier protein) synthase III, 3-oxoacyl-[acyl-carrier-protein] synthase 3, ... | | Authors: | Drees, S.L, Li, C, Prasetya, F, Saleem, M, Dreveny, I, Hennecke, U, Williams, P, Emsley, J, Fetzner, S. | | Deposit date: | 2015-09-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM.
J.Biol.Chem., 291, 2016
|
|
5E3A
 
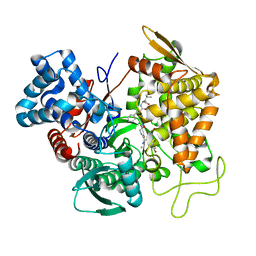 | | Structure of human DPP3 in complex with opioid peptide leu-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, Leu-enkephalin, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5FLH
 
 | | Free state of Ni-quercetinase | | Descriptor: | DIMETHYL SULFOXIDE, NICKEL (II) ION, NITRATE ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6FS3
 
 | | Phosphotriesterase PTE_A53_1 | | Descriptor: | (2~{S})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Bacterail Phosphotriesterase variant with high catalytic activity towards organophosphate nerve agents developed by use of structure-based design and molecular evolution
To Be Published
|
|
