5BUX
 
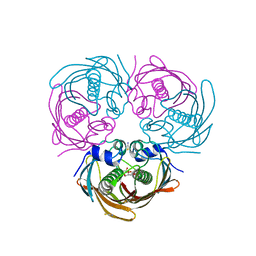 | |
3BCV
 
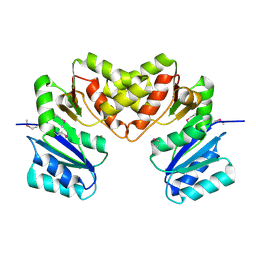 | |
3BGA
 
 | |
2GVC
 
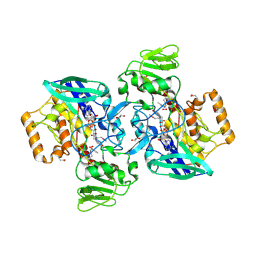 | | Crystal structure of flavin-containing monooxygenase (FMO)from S.pombe and substrate (methimazole) complex | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3C4R
 
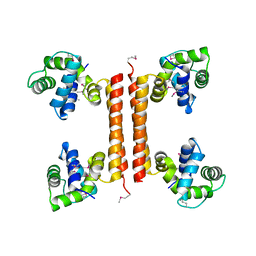 | | Crystal structure of an uncharacterized protein encoded by cryptic prophage | | Descriptor: | Uncharacterized protein | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, Ozyurt, S, Luz, J, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-19 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized protein encoded by cryptic prophage.
To be Published
|
|
2A9F
 
 | |
3C8A
 
 | |
3BQT
 
 | |
3CBR
 
 | | Crystal structure of human Transthyretin (TTR) at pH3.5 | | Descriptor: | Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Snee, W.C, Kelly, J.W, C Sacchettini, J. | | Deposit date: | 2008-02-22 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
3DEB
 
 | |
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
2I3O
 
 | |
3LXT
 
 | |
3NIV
 
 | |
2MI1
 
 | |
3NZG
 
 | | Crystal structure of a putative racemase with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative racemase | | Authors: | Eswaramoorthy, S, Raparia, E, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-07-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative racemase with Mg ion
TO BE PUBLISHED
|
|
2IMR
 
 | |
1YIR
 
 | |
2I6E
 
 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
2I76
 
 | | Crystal structure of protein TM1727 from Thermotoga maritima | | Descriptor: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
3M3M
 
 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3FIE
 
 | |
1YXW
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | GLUTAMIC ACID, TRYPTOPHAN, TYROSINE, ... | | Authors: | Jayaraman, S, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
1ZCC
 
 | | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens str.C58 | | Descriptor: | ACETATE ION, SULFATE ION, glycerophosphodiester phosphodiesterase | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens by SAD with a large asymmetric unit.
Proteins, 65, 2006
|
|
1YYN
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin | | Authors: | Seetharaman, J, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
