2PBZ
 
 | |
3NYW
 
 | |
2QVC
 
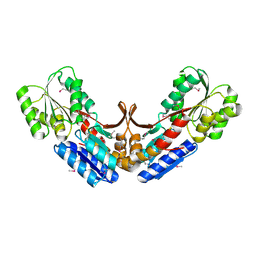 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | Descriptor: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2QZJ
 
 | |
3T81
 
 | |
2AFA
 
 | |
3PAN
 
 | |
3SMD
 
 | |
3PAO
 
 | |
3PU6
 
 | |
3Q0H
 
 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) | | Descriptor: | T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT)
To be published
|
|
3SPX
 
 | | Crystal structure of O-Acetyl Serine Sulfhydrylase from Leishmania donovani | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, O-acetyl serine sulfhydrylase, ... | | Authors: | Raj, I, Gourinath, S. | | Deposit date: | 2011-07-04 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The narrow active-site cleft of O-acetylserine sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-terminal sequences
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3Q1Y
 
 | |
2QVH
 
 | |
3T4P
 
 | |
3N8N
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 6 | | Descriptor: | (1R,4R,5R)-3-(tert-butylcarbamoyl)-1,4,5-trihydroxycyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3BQX
 
 | |
3N7A
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3JQX
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen binding domain 3 at 2.2 Angstrom resolution in the presence of calcium and cadmium | | Descriptor: | CADMIUM ION, CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Bauer, R, Matsushita, O. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3NAS
 
 | |
3CYG
 
 | |
3JQW
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen-binding domain 3 at 2 Angstrom resolution in presence of calcium | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Matsushita, O, Bauer, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3PBM
 
 | |
1YBF
 
 | |
2RDY
 
 | |
