5KBG
 
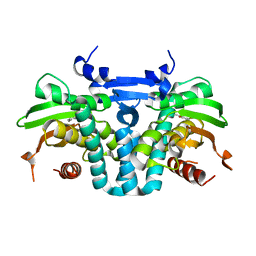 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | Descriptor: | MopR, ZINC ION, o-cresol | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBE
 
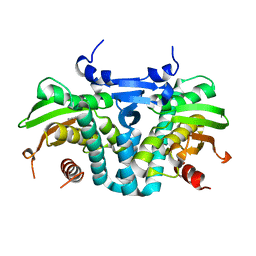 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH PHENOL | | Descriptor: | MopR, PHENOL, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBH
 
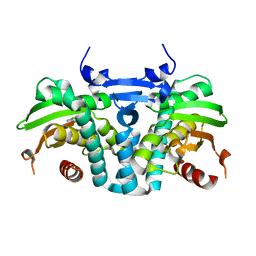 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH 3-CHLORO-PHENOL | | Descriptor: | 3-CHLOROPHENOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
4XWO
 
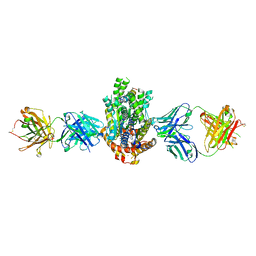 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4U5Q
 
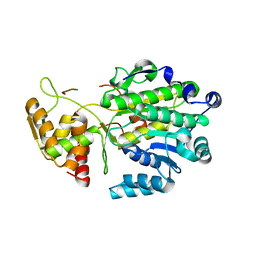 | | High resolution crystal structure of reductase (R) domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Peptide synthetase | | Authors: | Patel, K.D, Haque, A.S, Priyadarshan, K, Sankaranarayanan, R. | | Deposit date: | 2014-07-25 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | High resolution structure of R-domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis
To Be Published
|
|
6MZT
 
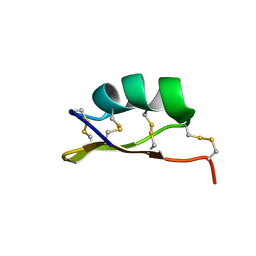 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | Descriptor: | Potassium channel toxin alpha-KTx 6.21 | | Authors: | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | Deposit date: | 2018-11-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
6Y3D
 
 | | X-ray structure of thermophilic C-phycocyanin from Galdiera phlegrea | | Descriptor: | ACETATE ION, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Ferraro, G, Lucignano, R, Marseglia, A, Merlino, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of C-phycocyanin from Galdieria phlegrea: Determinants of thermostability and comparison with a C-phycocyanin in the entire phycobilisome.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
4TSS
 
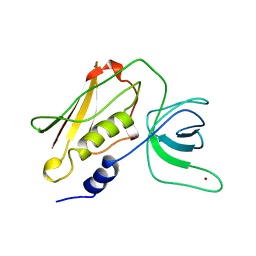 | | TOXIC SHOCK SYNDROME TOXIN-1: TETRAGONAL P4(1)2(1)2 CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1, ZINC ION | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
4XTR
 
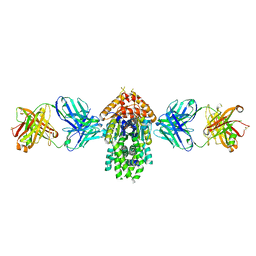 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
6WMP
 
 | | F. tularensis RNAPs70-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA T-strand, DNA-directed RNA polymerase subunit alpha 1, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
4RMP
 
 | | Crystal structure of allophycocyanin from marine cyanobacterium Phormidium sp. A09DM | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Kumar, V, Gupta, G.D, Sonani, R.R, Madamwar, D. | | Deposit date: | 2014-10-22 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of Allophycocyanin from Marine Cyanobacterium Phormidium sp. A09DM.
Plos One, 10, 2015
|
|
6OF0
 
 | | Structural basis for multidrug recognition and antimicrobial resistance by MTRR, an efflux pump regulator from Neisseria Gonorrhoeae | | Descriptor: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Beggs, G.A, Kumaraswami, M, Shafer, W, Brennan, R.G. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and In Vivo Characterization of MtrR-Mediated Resistance to Innate Antimicrobials by the Human Pathogen Neisseria gonorrhoeae .
J.Bacteriol., 201, 2019
|
|
5XKP
 
 | | Crystal structure of Msmeg3575 in complex with 5-azacytosine | | Descriptor: | 6-amino-1,3,5-triazin-2(1H)-one, ACETATE ION, CMP/dCMP deaminase, ... | | Authors: | Gaded, V.M, Anand, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
6IFX
 
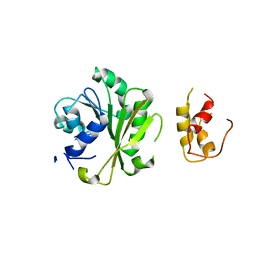 | |
6JT9
 
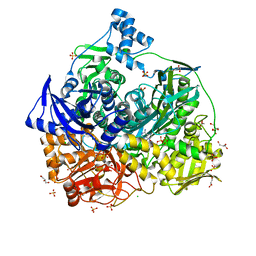 | | Crystal Structure of D464A mutant of FGAM Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6IFS
 
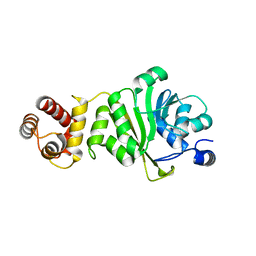 | | KsgA from Bacillus subtilis 168 | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Bhujbalrao, R, Anand, R. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance.
J. Am. Chem. Soc., 141, 2019
|
|
6IFV
 
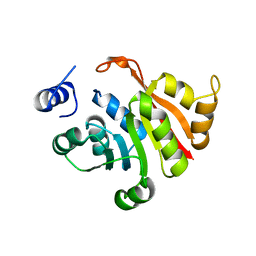 | | C-terminal truncated KsgA from Bacillus subtilis 168 | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Bhujbalrao, R, Anand, R. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance.
J. Am. Chem. Soc., 141, 2019
|
|
5XAZ
 
 | |
5XAY
 
 | |
5XKR
 
 | | Crystal structure of Msmeg3575 in complex with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ACETATE ION, CMP/dCMP deaminase, ... | | Authors: | Gaded, V.M, Anand, R. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
6JT8
 
 | | Crystal structure of 450-451_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6WEG
 
 | |
7V2N
 
 | | T.thermophilus 30S ribosome with KsgA, class K2 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2O
 
 | | T.thermophilus 30S ribosome with KsgA, class K4 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
