4KWB
 
 | |
3RST
 
 | |
5OWF
 
 | | Structure of a LAO-binding protein mutant with glutamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | Authors: | Shanmugaratnam, S, Banda-Vazquez, J, Sosa-Peinado, A, Hocker, B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Redesign of LAOBP to bind novel l-amino acid ligands.
Protein Sci., 27, 2018
|
|
7QSP
 
 | | Permutated C-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7QSQ
 
 | | Permutated N-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein, ... | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1QBM
 
 | |
7Q1R
 
 | | A de novo designed homo-dimeric antiparallel coiled coil apCC-Di | | Descriptor: | ETHANOL, SODIUM ION, apCC-Di | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
5T3M
 
 | |
7Q1T
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB | | Descriptor: | N-PROPANOL, SULFATE ION, apCC-Di-A, ... | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
6KJL
 
 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
7A8S
 
 | | de novo designed ba8-barrel sTIM11 with an alpha-helical extension | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, sTIM11_h3 | | Authors: | Shanmugaratnam, S, Wiese, J.G, Hocker, B. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Extension of a de novo TIM barrel with a rationally designed secondary structure element.
Protein Sci., 30, 2021
|
|
1K28
 
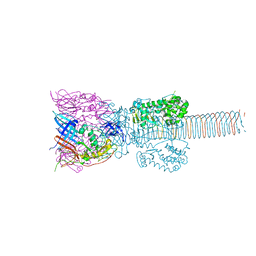 | | The Structure of the Bacteriophage T4 Cell-Puncturing Device | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP27, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kanamaru, S, Leiman, P.G, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Arisaka, F, Rossmann, M.G. | | Deposit date: | 2001-09-26 | | Release date: | 2002-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the cell-puncturing device of bacteriophage T4.
Nature, 415, 2002
|
|
1QBL
 
 | |
7N6T
 
 | |
7N6V
 
 | |
7N6X
 
 | |
7Q1S
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB_var | | Descriptor: | 1,2-ETHANEDIOL, apCC-Di-A_var, apCC-Di-B_var | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
4L1D
 
 | | Voltage-gated sodium channel beta3 subunit Ig domain | | Descriptor: | Sodium channel subunit beta-3 | | Authors: | Namadurai, S, Weimhofer, M, Rajappa, R, Stott, K, Klingauf, J, Chirgadze, D.Y, Jackson, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Molecular Imaging of the Nav Channel beta 3 Subunit Indicates a Trimeric Assembly.
J.Biol.Chem., 289, 2014
|
|
7OYZ
 
 | | E.coli's putrescine receptor variant PotF/D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYU
 
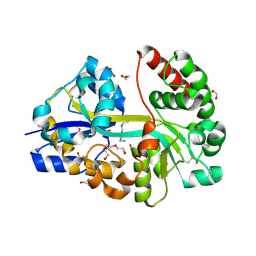 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S F88Y in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7Q1Q
 
 | | De novo designed homo-dimeric antiparallel helices Homomer-S | | Descriptor: | ACETATE ION, Homomer-S | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
1SPG
 
 | |
3RYL
 
 | |
7CN6
 
 | | T4 phage spackle protein gp61.3 | | Descriptor: | CALCIUM ION, Protein spackle | | Authors: | Kanamaru, S, Leiman, P.G. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of the T4 Spackle Protein Gp61.3.
Viruses, 12, 2020
|
|
7CN7
 
 | |
