3F01
 
 | |
6FAL
 
 | |
6F7F
 
 | |
6F7G
 
 | |
8DD6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant in Complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, DIMETHYL SULFOXIDE, ORF1a polyprotein | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-17 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
6F9K
 
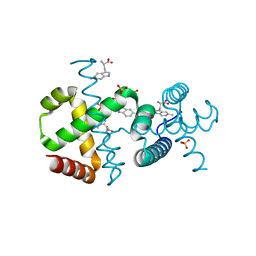 | |
7BPN
 
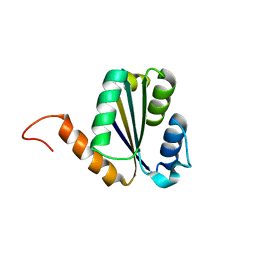 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
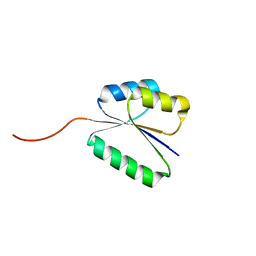 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
3F00
 
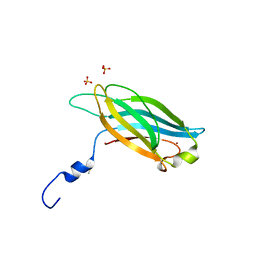 | |
1PDF
 
 | | Fitting of gp11 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 baseplate-tail tube complex | | Descriptor: | Baseplate structural protein Gp11 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDL
 
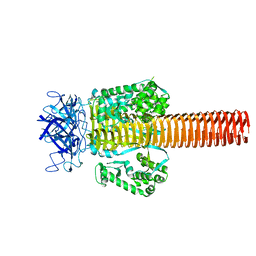 | | Fitting of gp5 in the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Tail-associated lysozyme | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
3WIT
 
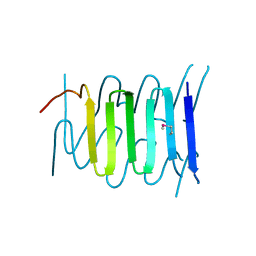 | |
1PDM
 
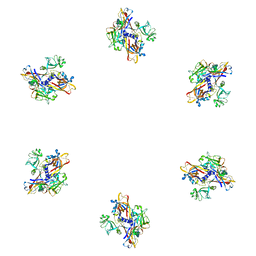 | | Fitting of gp8 structure into the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp8 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
7BQE
 
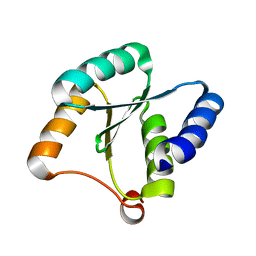 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | Descriptor: | NF6 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
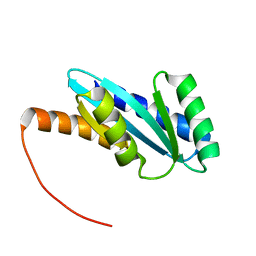 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
2HUG
 
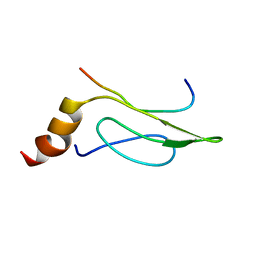 | | 3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplast, Signal recognition particle 54 kDa protein | | Authors: | Kathir, K.M, Vaithiyalingam, S, Henry, R, Thallapuranam, S.K.K. | | Deposit date: | 2006-07-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43.
J.Mol.Biol., 381, 2008
|
|
1NO4
 
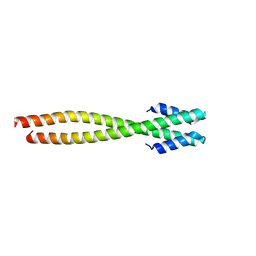 | | Crystal Structure of the pre-assembly scaffolding protein gp7 from the double-stranded DNA bacteriophage phi29 | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.A.L, McMurray, C.T, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2003-01-15 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
7BPL
 
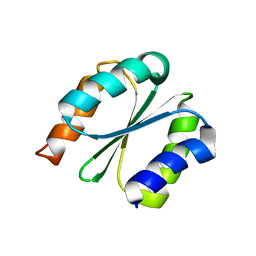 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | Descriptor: | NF1 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPP
 
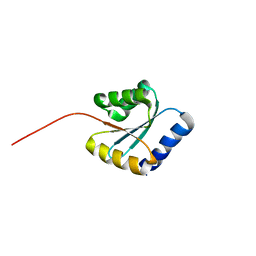 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | Descriptor: | NF5 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
1PDJ
 
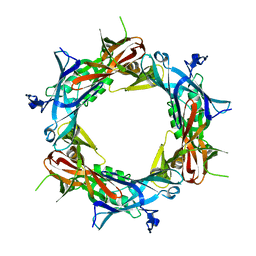 | | Fitting of gp27 into cryoEM reconstruction of bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp27 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDP
 
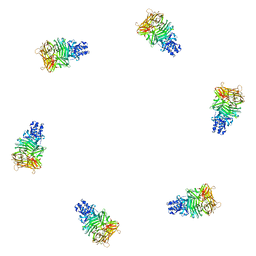 | | Fitting of gp9 structure into the bacteriophage T4 baseplate cryoEM reconstruction | | Descriptor: | Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1NOH
 
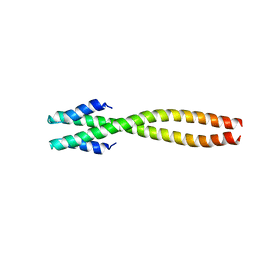 | | The structure of bacteriophage phi29 scaffolding protein gp7 after prohead assembly | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.L, McMurray, C.T, L Anderson, D, Rossmann, M.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
8R8O
 
 | | Hallucinated de novo TIM barrel with three helical extensions - HalluTIM3-1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Beck, J, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Diversifying de novo TIM barrels by hallucination.
Protein Sci., 33, 2024
|
|
