6KCA
 
 | |
6JXP
 
 | |
6KD1
 
 | |
7CJZ
 
 | |
7CK0
 
 | |
7CVK
 
 | |
7CVJ
 
 | |
7CVL
 
 | |
7CJO
 
 | | Crystal structure of metal-bound state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7CJP
 
 | | Crystal structure of metal-free state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7BVL
 
 | |
7BVO
 
 | |
7BVM
 
 | |
7BVN
 
 | |
7CVM
 
 | |
6LOF
 
 | | Crystal structure of ZsYellow soaked by Cu2+ | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Nam, K.H. | | Deposit date: | 2020-01-05 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Spectroscopic and Structural Analysis of Cu 2+ -Induced Fluorescence Quenching of ZsYellow.
Biosensors (Basel), 10, 2020
|
|
6LZO
 
 | | Thermolysin with 1,10-phenanthroline | | Descriptor: | 1,10-PHENANTHROLINE, CALCIUM ION, Thermolysin | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
6LZN
 
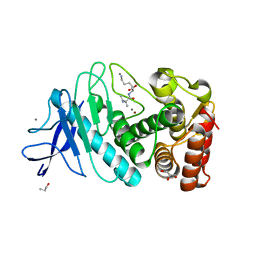 | | Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
6LL2
 
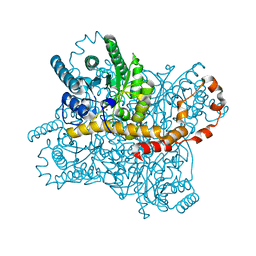 | |
6LL3
 
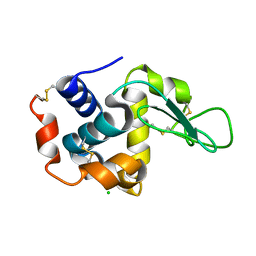 | |
8WGK
 
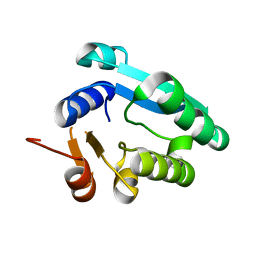 | |
8WDG
 
 | |
8WFU
 
 | |
8WDI
 
 | |
8WFW
 
 | |
