2BGZ
 
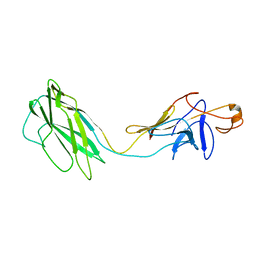 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5AX2
 
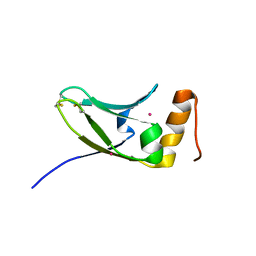 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
2D4Y
 
 | |
5H53
 
 | |
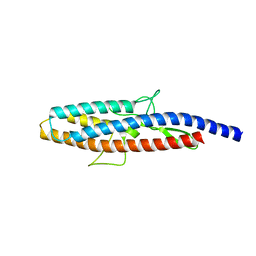
2D4X
 
 | |
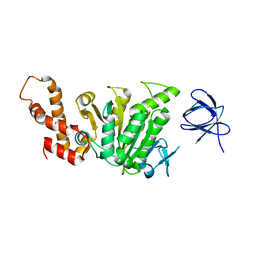
2DPY
 
 | |
6KN7
 
 | | Structure of human cardiac thin filament in the calcium free state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
6KN8
 
 | | Structure of human cardiac thin filament in the calcium bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
6K9Q
 
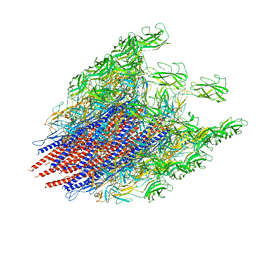 | | Structure of the native supercoiled hook as a universal joint | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Kato, T, Miyata, T, Makino, F, Horvath, P, Namba, K. | | Deposit date: | 2019-06-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the native supercoiled flagellar hook as a universal joint.
Nat Commun, 10, 2019
|
|
5WRH
 
 | |
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
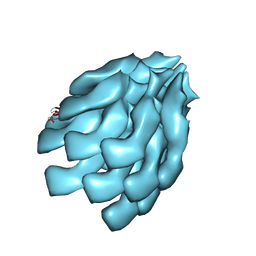
6KFK
 
 | |
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
7VW6
 
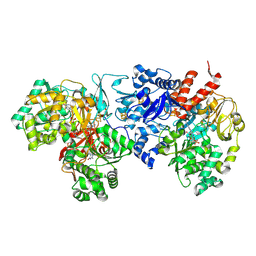 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
3VJP
 
 | |
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
7W2J
 
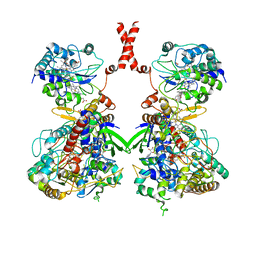 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
4A6J
 
 | | Structural model of ParM filament based on CryoEM map | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Gayathri, P, Fujii, T, Moller-Jensen, J, Van Den Ent, F, Namba, K, Lowe, J. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
3VKI
 
 | |
7WT6
 
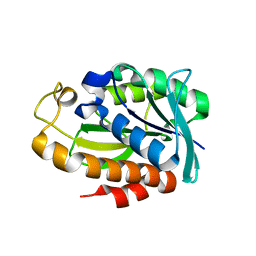 | |
3TEE
 
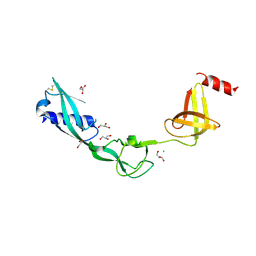 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
3AJW
 
 | | Structure of FliJ, a soluble component of flagellar type III export apparatus | | Descriptor: | Flagellar fliJ protein, MERCURY (II) ION | | Authors: | Imada, K, Ibuki, T, Minamino, T, Namba, K. | | Deposit date: | 2010-06-23 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases
Nat.Struct.Mol.Biol., 18, 2011
|
|
2D4V
 
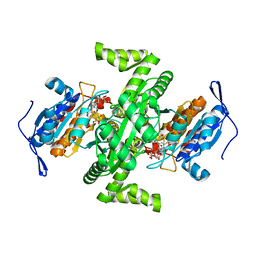 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | Descriptor: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | Authors: | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
3A7M
 
 | | Structure of FliT, the flagellar type III chaperone for FliD | | Descriptor: | Flagellar protein fliT | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
