4KOC
 
 | |
4KO7
 
 | |
4KO5
 
 | | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F) | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Inampudi, K.K, Wang, Y, Meng, W, Tobin, P.H, Wilson, C.J. | | Deposit date: | 2013-05-11 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F)
To be Published
|
|
4KOB
 
 | |
4KO9
 
 | |
7VEW
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
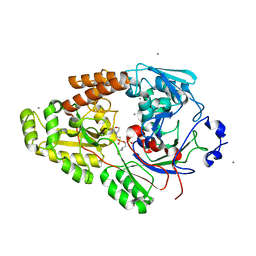 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
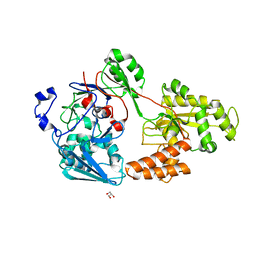 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
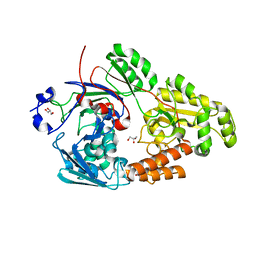 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
1IKL
 
 | |
1IKM
 
 | |
1G91
 
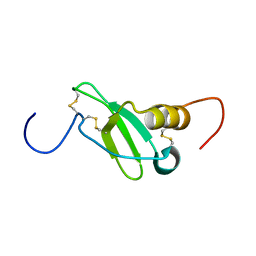 | | SOLUTION STRUCTURE OF MYELOID PROGENITOR INHIBITORY FACTOR-1 (MPIF-1) | | Descriptor: | MYELOID PROGENITOR INHIBITORY FACTOR-1 | | Authors: | Rajarathnam, K, Li, Y, Rohrer, T, Gentz, R. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-07 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine.
J.Biol.Chem., 276, 2001
|
|
5HXY
 
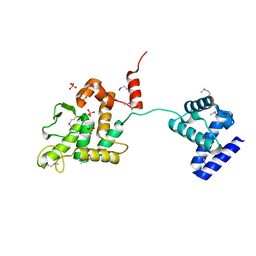 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
7R21
 
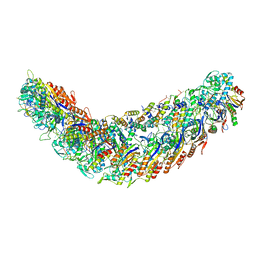 | | elongated Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | Cas11a, Cas7a, CrRNA (62-MER), ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots for an atypic type I CRISPR-Cas system
To Be Published
|
|
7R2K
 
 | | elongated Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Terns, M, Stahlberg, H, Ke, A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots for an atypic type I CRISPR-Cas system
To Be Published
|
|
8Q2B
 
 | | E. coli Adenylate Kinase variant D158A (AK D158A) showing significant changes to the stacking of catalytic arginine side chains | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Sauer, U.H, Wolf-Watz, M, Nam, K. | | Deposit date: | 2023-08-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.
J.Chem.Inf.Model., 64, 2024
|
|
7APU
 
 | | Structure of Adenylate kinase from Escherichia coli in complex with two ADP molecules refined at 1.36 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, SODIUM ION | | Authors: | Grundstom, C, Wolf-Watz, M, Nam, K, Sauer, U.H. | | Deposit date: | 2020-10-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions.
Biochemistry, 60, 2021
|
|
3II1
 
 | |
3H18
 
 | | Crystal structure of EstE5-PMSF (II) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3H17
 
 | | Crystal structure of EstE5-PMSF (I) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3K6K
 
 | |
3G6N
 
 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3GVY
 
 | |
