1UI8
 
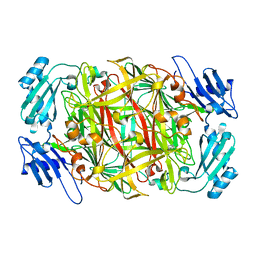 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
3HEC
 
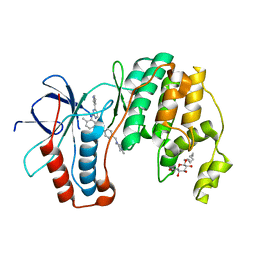 | | P38 in complex with Imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
3HEG
 
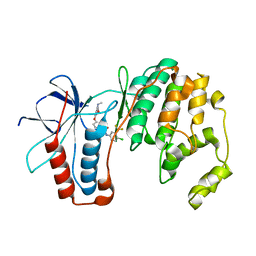 | | P38 in complex with Sorafenib | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Mitogen-activated protein kinase 14 | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
3TEE
 
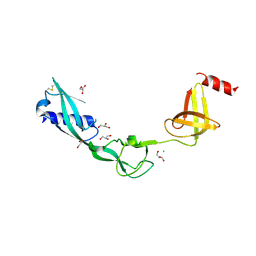 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
3VJP
 
 | |
3VKI
 
 | |
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
3TT3
 
 | |
3TT1
 
 | |
3TU0
 
 | |
3O8U
 
 | |
3O8P
 
 | | Conformational plasticity of p38 MAP kinase DFG motif mutants in response to inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, Mitogen-activated protein kinase 14, ... | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational plasticity of p38 MAP kinase DFG motif mutants in response to inhibitor binding
To be Published
|
|
3P5K
 
 | | P38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(1,1-dioxidothiomorpholin-4-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H. | | Deposit date: | 2010-10-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3O8T
 
 | |
3OC1
 
 | | Conformational plasticity of p38 MAP kinase DFG motif mutants in response to inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, Mitogen-activated protein kinase 14 | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Conformational plasticity of p38 MAP kinase DFG motif mutants in response to inhibitor binding
To be Published
|
|
3OBG
 
 | |
3OBJ
 
 | |
3P7K
 
 | |
4IOV
 
 | | The structure of AAVrh32.33, a Novel Gene Delivery Vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein VP1 | | Authors: | Mikalism, K, Nam, H.-J, Van vilet, K, Vandenberghe, L.H, Mays, L.E, McKenna, R, Wilson, J, Agbandje-McKenna, M. | | Deposit date: | 2013-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of AAVrh32.33, a novel gene delivery vector.
J.Struct.Biol., 186, 2014
|
|
2JMK
 
 | | Solution structure of ta0956 | | Descriptor: | Hypothetical protein Ta0956 | | Authors: | Koo, B, Jung, J, Jung, H, Nam, H, Kim, Y, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-11-20 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical novel-fold protein TA0956 from Thermoplasma acidophilum
Proteins, 69, 2007
|
|
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
5KAZ
 
 | | Human SH2D1B structure | | Descriptor: | SH2 domain-containing protein 1B, SULFATE ION | | Authors: | Taha, M, Nezerwa, E, Nam, H.-J. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray Crystallographic Structure of Human EAT2 (SH2D1B).
Protein Pept. Lett., 23, 2016
|
|
5BQD
 
 | | Crystal Structure of TBX5 (1-239) Dimer | | Descriptor: | MAGNESIUM ION, T-box transcription factor TBX5 | | Authors: | Pradhan, L, Gopal, S, Patel, A, Kasahara, H, Nam, H.J. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
1Z1C
 
 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute virus of Mice | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*AP*CP*AP*CP*CP*AP*AP*AP*A)-3', 5'-D(*AP*TP*CP*CP*TP*CP*TP*AP*TP*CP*AP*C)-3', ... | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
1Z14
 
 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute Virus of Mice | | Descriptor: | VP2 | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
