1OGZ
 
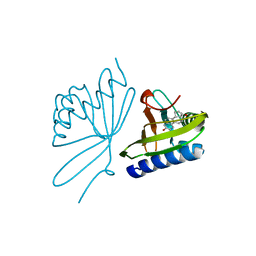 | | Crystal Structure Of 5-3-Ketosteroid Isomerase Mutants P39A Complexed With Equilenin From Pseudomonas Testosteroni | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Nam, G.H, Cha, S.-S, Yun, Y.S, Oh, Y.H, Hong, B.H, Lee, H.-S, Choi, K.Y. | | Deposit date: | 2003-05-20 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Conserved Cis-Pro39 Residue Plays a Crucial Role in the Proper Positioning of the Catalytic Base Asp38 in Ketosteroid Isomerase from Comamonas Testosteroni.
Biochem.J., 375, 2003
|
|
4LLA
 
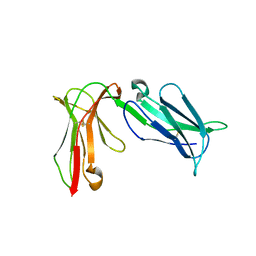 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4LL9
 
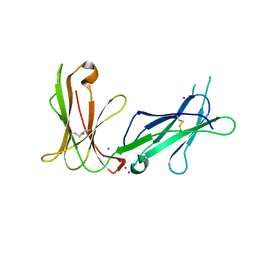 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
3P2T
 
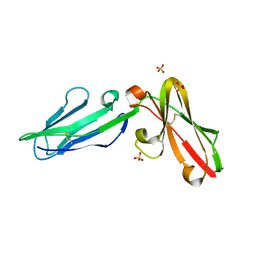 | | Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, SULFATE ION | | Authors: | Chen, Y, Nam, G, Cheng, H, Zhang, J.H, Willcox, B.E, Gao, G.F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance
J.Biol.Chem., 286, 2011
|
|
1W6Y
 
 | | crystal structure of a mutant W92A in ketosteroid isomerase (KSI) from Pseudomonas putida biotype B | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Yun, Y.S, Nam, G.H, Kim, Y.-G, Oh, B.-H, Choi, K.Y. | | Deposit date: | 2004-08-25 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Exterior Hydrophobic Cluster Contributes to Conformational Stability and Steroid Binding in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
FEBS J., 272, 2005
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1K41
 
 | | Crystal structure of KSI Y57S mutant | | Descriptor: | Ketosteroid Isomerase | | Authors: | Cha, S.S, Oh, B.H, Nam, G.H, Jang, D.S, Lee, T.H, Choi, K.Y. | | Deposit date: | 2001-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Maintenance of alpha-helical structures by phenyl rings in the active-site tyrosine triad contributes to catalysis and stability of ketosteroid isomerase from Pseudomonas putida biotype B
Biochemistry, 40, 2001
|
|
1DMN
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMQ
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
2LFT
 
 | | Human prion protein with E219K protective polymorphism | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giacin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-07-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
2LSB
 
 | | Solution-state NMR structure of the human prion protein | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giancin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
2LV1
 
 | | Solution-state NMR structure of prion protein mutant V210I at neutral pH | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Legname, G, Plavec, J. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structural Rearrangements at Physiological pH: Nuclear Magnetic Resonance Insights from the V210I Human Prion Protein Mutant.
Biochemistry, 51, 2012
|
|
2LEJ
 
 | | human prion protein mutant HuPrP(90-231, M129, V210I) | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Raspadori, A, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Molecular Basis of Inherited Prion Diseases: NMR Structure of the Human Prion Protein with V210I Mutation.
J.Mol.Biol., 412, 2011
|
|
2KUN
 
 | | Three dimensional structure of HuPrP(90-231 M129 Q212P) | | Descriptor: | Major prion protein | | Authors: | Ilc, G, Giachin, G, Jaremko, M, Jaremko, L, Zhukov, I, Plavec, J, Legname, G, Benetti, F. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human prion protein with the pathological Q212P mutation reveals unique structural features.
Plos One, 5, 2010
|
|
5LKN
 
 | | NMR solution structure of human FNIII domain 2 of NCAM | | Descriptor: | Neural cell adhesion molecule 1 | | Authors: | Slapsak, U, Salzano, G, Amin, L, Abskharon, R.N.N, Ilc, G, Zupancic, B, Biljan, I, Plavec, J, Giachin, G, Legname, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain.
J.Biol.Chem., 291, 2016
|
|
4N9O
 
 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
468D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
470D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
469D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
471D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
1WV6
 
 | | X-ray structure of the A-decamer GCGTATACGC with a single 2'-O-butyl thymine in place of T6, Sr-form | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2BT)P*AP*CP*GP*C)-3', STRONTIUM ION | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharam, M. | | Deposit date: | 2004-12-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2'-O-ribonucleic acid modifications.
Biochemistry, 44, 2005
|
|
1WV5
 
 | | X-ray structure of the A-decamer GCGTATACGC with a single 2'-o-butyl thymine in place of T6, Mg-form | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2BT)P*AP*CP*GP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharam, M. | | Deposit date: | 2004-12-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2'-O-ribonucleic acid modifications.
Biochemistry, 44, 2005
|
|
1Y8L
 
 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(trifluoro)ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(TFE)P*AP*CP*GP*C)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
