7YF6
 
 | | Crystal structure of HIV-1 protease in complex with macrocyclic peptide | | Descriptor: | MAGNESIUM ION, Macrocyclic Peptide, Protease | | Authors: | Kusumoto, Y, Sato, S, Yamada, T, Kozono, I, Nakata, Z, Asada, N, Mitsuki, S, Wakasa-Morimoto, C, Tohru, M, Watanabe, A, Hayashi, K, Mikamiyama, H. | | Deposit date: | 2022-07-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Highly Potent and Oral Macrocyclic Peptides as a HIV-1 Protease Inhibitor: mRNA Display-Derived Hit-to-Lead Optimization.
Acs Med.Chem.Lett., 13, 2022
|
|
8YTD
 
 | | Crystal Structure of TrkA D5 domain in complex with two different macrocyclic peptides | | Descriptor: | 1,2-ETHANEDIOL, High affinity nerve growth factor receptor, Macrocyclic Peptide | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
8YTE
 
 | | Crystal Structure of TrkA D5 domain in complex with macrocyclic peptide | | Descriptor: | 1,2-ETHANEDIOL, AMINOMETHYLAMIDE, High affinity nerve growth factor receptor, ... | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
7WBS
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WCQ
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
3WSY
 
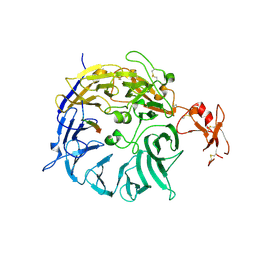 | | SorLA Vps10p domain in complex with its own propeptide fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor, peptide from Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSZ
 
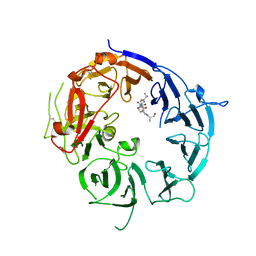 | | SorLA Vps10p domain in complex with Abeta-derived peptide | | Descriptor: | 10-mer peptide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSX
 
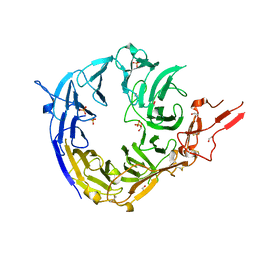 | | SorLA Vps10p domain in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
