3W20
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3W0R
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N202A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W21
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3W0Q
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N203A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0P
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (D198A), ternary complex with ADP and hygromycin B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0N
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0O
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with ADP and hygromycin B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0S
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, MAGNESIUM ION, ... | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0M
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3WKR
 
 | | Crystal structure of the SepCysS-SepCysE complex from Methanocaldococcus jannaschii | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3W9W
 
 | | Crystal structure of DING protein | | Descriptor: | DING protein, GLYCEROL, PHOSPHATE ION | | Authors: | Gai, Z.Q, Nakamura, A, Tanaka, Y, Hirano, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure analysis, overexpression and refolding behaviour of a DING protein with single mutation.
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3WKS
 
 | | Crystal structure of the SepCysS-SepCysE N-terminal domain complex from | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.029 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3W9V
 
 | | Crystal structure of refolded DING protein | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphate-binding protein | | Authors: | Gai, Z.Q, Nakamura, A, Tanaka, Y, Hirano, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.031 Å) | | Cite: | Crystal structure analysis, overexpression and refolding behaviour of a DING protein with single mutation.
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5ZD4
 
 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
5ZL9
 
 | |
7Y8U
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8V
 
 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8X
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
5GWT
 
 | | 4-hydroxyisoleucine dehydrogenase mutant complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
5GWS
 
 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
5GWR
 
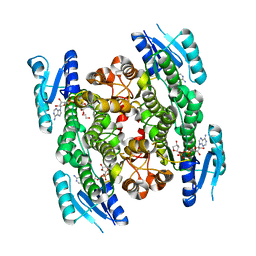 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers
Sci Rep, 7, 2017
|
|
5H3E
 
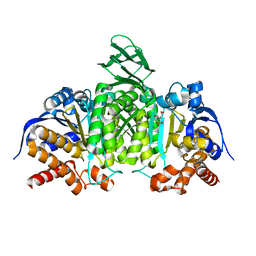 | | Crystal structure of mouse isocitrate dehydrogenases 2 K256Q mutant complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
5H3F
 
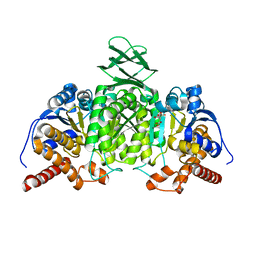 | | Crystal structure of mouse isocitrate dehydrogenases 2 complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
1PAM
 
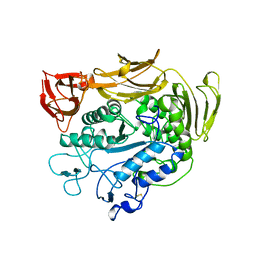 | | CYCLODEXTRIN GLUCANOTRANSFERASE | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Harata, K, Haga, K, Nakamura, A, Aoyagi, M, Yamane, K. | | Deposit date: | 1996-07-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011. Comparison of two independent molecules at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6KTK
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
