5YAQ
 
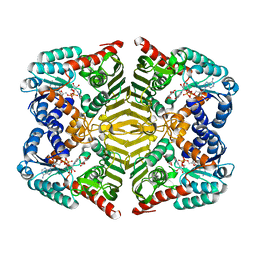 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with scyllo-inosose | | 分子名称: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 著者 | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2017-09-01 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAB
 
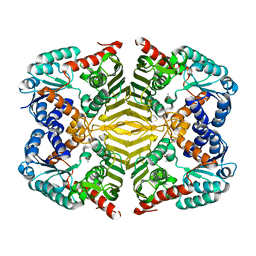 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 分子名称: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 著者 | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2017-08-31 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YA8
 
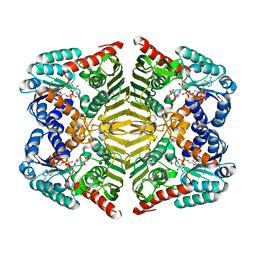 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with myo-inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 著者 | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2017-08-31 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAP
 
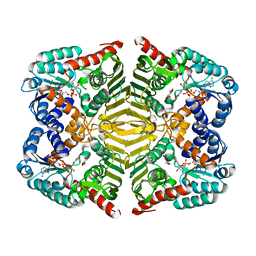 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with L-glucono-1,5-lactone | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | 著者 | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2017-09-01 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5Z7X
 
 | | Crystal structure of Striga hermonthica HTL4 (ShHTL4) | | 分子名称: | 1,2-ETHANEDIOL, Hyposensitive to light 4, MAGNESIUM ION | | 著者 | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.055 Å) | | 主引用文献 | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
7Y8X
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | 著者 | Ishida, K, Nakamura, A, Kojima, S. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8U
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | 分子名称: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | 著者 | Ishida, K, Nakamura, A, Kojima, S. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8V
 
 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | 分子名称: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | 著者 | Ishida, K, Nakamura, A, Kojima, S. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | 分子名称: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | 登録日 | 2019-04-10 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
5YP4
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
6IY9
 
 | | Crystal structure of aminoglycoside 7"-phoshotransferase-Ia (APH(7")-Ia/HYG) from Streptomyces hygroscopicus complexed with hygromycin B | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, HYGROMYCIN B VARIANT, ... | | 著者 | Takenoya, M, Shimamura, T, Yamanaka, R, Adachi, Y, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2018-12-14 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5YP2
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | 分子名称: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
6JTC
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | 分子名称: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | 登録日 | 2019-04-10 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5ZD4
 
 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | 著者 | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | 登録日 | 2018-02-22 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
5XCY
 
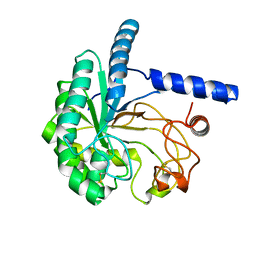 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | 分子名称: | Glucanase | | 著者 | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | 登録日 | 2017-03-24 | | 公開日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (1.199 Å) | | 主引用文献 | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1IJ6
 
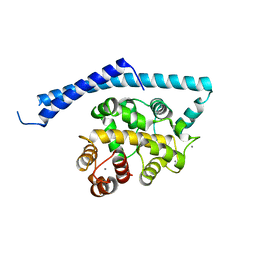 | | CA2+-BOUND STRUCTURE OF MULTIDOMAIN EF-HAND PROTEIN, CBP40, FROM TRUE SLIME MOLD | | 分子名称: | CALCIUM ION, PLASMODIAL SPECIFIC LAV1-2 PROTEIN | | 著者 | Iwasaki, W, Sasaki, H, Nakamura, A, Kohama, K, Tanokura, M. | | 登録日 | 2001-04-25 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Metal-Free and Ca(2+)-Bound Structures of a Multidomain EF-Hand Protein, CBP40, from the Lower Eukaryote Physarum polycephalum
Structure, 11, 2003
|
|
5Y33
 
 | |
5ZL9
 
 | |
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | 登録日 | 2019-08-28 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
7DKD
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | 分子名称: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | 分子名称: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | 分子名称: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | 著者 | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-12-14 | | 公開日 | 2006-05-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
7DKC
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | 分子名称: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
