2WR8
 
 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
2XG6
 
 | |
2C4U
 
 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2X0Q
 
 | |
2XE2
 
 | |
2XE1
 
 | |
2C4T
 
 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with an inhibitor, an analogue of S- adenosyl methionine | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, CHLORIDE ION, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrates and Inhibitors of the Fluorinase from Streptomyces Cattleya
To be Published
|
|
5O78
 
 | |
5OUT
 
 | |
5NR2
 
 | |
5NC4
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with protochelin from Pseudomonas aeruginosa | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[(5~{S})-5-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-6-[4-[[2,3-bis(oxidanyl)phenyl]carbonylamino]butylamino]-6-oxidanylidene-hexyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2017-03-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
5O3U
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin F1 | | Descriptor: | Peptide cyclase 1, Putative presegetalin F1 | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5N0R
 
 | |
5N4I
 
 | |
5O3V
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin B1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Putative presegetalin B1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O9C
 
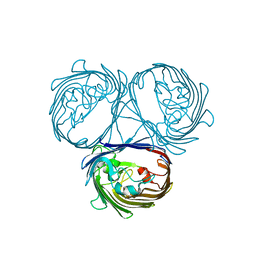 | |
5N4D
 
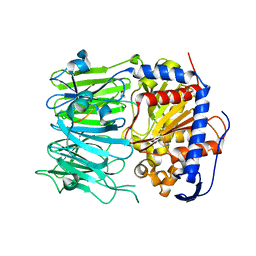 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - D661A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4C
 
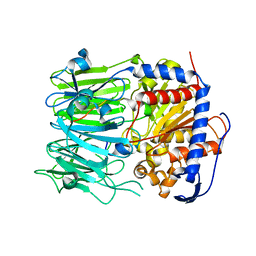 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5O3W
 
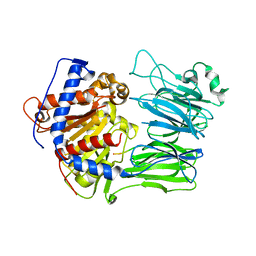 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin A1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
8B72
 
 | |
4BMK
 
 | | Serine Palmitoyltransferase K265A from S. paucimobilis with bound PLP- Myriocin Aldimine | | Descriptor: | Decarboxylated Myriocin, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wadsworth, J.M, Clarke, D.J, McMahon, S.A, Beattie, A.E, Lowther, J, Dunn, T.M, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin.
J.Am.Chem.Soc., 135, 2013
|
|
5OUF
 
 | |
4AI4
 
 | |
5N0T
 
 | |
5N4F
 
 | | Prolyl oligopeptidase B from Galerina marginata - apo protein | | Descriptor: | GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
