6C28
 
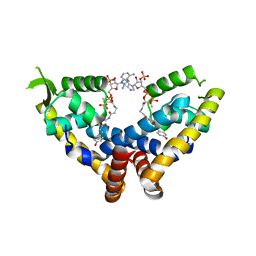 | | Transcriptional repressor, CouR, bound to p-coumaroyl-CoA | | Descriptor: | Transcriptional regulator, MarR family, p-coumaroyl-CoA | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-07 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6CGQ
 
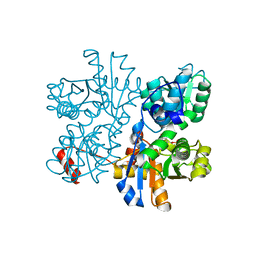 | | Threonine synthase from Bacillus subtilis ATCC 6633 with PLP and PLP-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.
ACS Chem. Biol., 14, 2019
|
|
6D6N
 
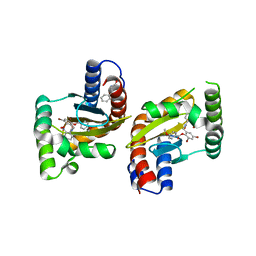 | | The structure of ligand binding domain of LasR in complex with TP-1 homolog, compound 16 | | Descriptor: | 2,4-dibromo-6-{[(2-nitrobenzene-1-carbonyl)amino]methyl}phenyl 4-methoxybenzoate, PHENYLALANINE, Transcriptional activator protein LasR | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop "Out" Conformation for LasR.
Cell Chem Biol, 25, 2018
|
|
6C0G
 
 | |
6D6M
 
 | |
6D6A
 
 | |
6C2S
 
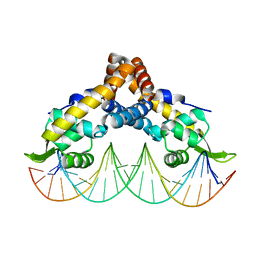 | | Transcriptional repressor, CouR, bound to a 23-mer DNA duplex | | Descriptor: | 23-mer, Transcriptional regulator, MarR family | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C8R
 
 | |
6C0Y
 
 | |
6C8S
 
 | | Loganic acid methyltransferase with SAH | | Descriptor: | Loganic acid O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Loganic Acid Methyltransferase: Insights into the Specificity of Methylation on an Iridoid Glycoside.
Chembiochem, 19, 2018
|
|
4IM7
 
 | | Crystal structure of fructuronate reductase (ydfI) from E. coli CFT073 (EFI TARGET EFI-506389) complexed with NADH and D-mannonate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-MANNONIC ACID, Hypothetical oxidoreductase ydfI, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-01-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of fructuronate reductase (ydfI) from E. coli CFT073 (EFI TARGET EFI-506389) complexed with NADH and D-mannonate
To be Published
|
|
4K8G
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G) | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Lukk, T, Wichelecki, D, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G)
To be Published
|
|
4MGS
 
 | | BiXyn10A CBM1 APO | | Descriptor: | Putative glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NEI
 
 | | Alg17c PL17 Family Alginate Lyase | | Descriptor: | ZINC ION, alginate lyase | | Authors: | Park, D.S, Nair, S.K. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
4QEC
 
 | | ElxO with NADP Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4Q85
 
 | | YcaO with Non-hydrolyzable ATP (AMPCPP) Bound | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4MWZ
 
 | |
4Q84
 
 | | Apo YcaO | | Descriptor: | MERCURY (II) ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4Q86
 
 | | YcaO with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4QED
 
 | | ElxO Y152F with NADPH Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4R9F
 
 | | CpMnBP1 with Mannobiose Bound | | Descriptor: | MBP1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4R9G
 
 | | CpMnBP1 with Mannotriose Bound | | Descriptor: | MBP1, ZINC ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4QPW
 
 | | BiXyn10A CBM1 with Xylohexaose Bound | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5U55
 
 | | Psf4 in complex with Mn2+ and (S)-2-HPP | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5U5D
 
 | | Psf4 in complex with Mn2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
