6C8S
 
 | | Loganic acid methyltransferase with SAH | | Descriptor: | Loganic acid O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Loganic Acid Methyltransferase: Insights into the Specificity of Methylation on an Iridoid Glycoside.
Chembiochem, 19, 2018
|
|
6C28
 
 | | Transcriptional repressor, CouR, bound to p-coumaroyl-CoA | | Descriptor: | Transcriptional regulator, MarR family, p-coumaroyl-CoA | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-07 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C8R
 
 | |
5BSV
 
 | |
5BSW
 
 | |
5BSU
 
 | | Crystal structure of 4-coumarate:CoA ligase complexed with caffeoyl adenylate | | Descriptor: | 4-coumarate--CoA ligase 2, 5'-O-[(R)-{[(2E)-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)prop-2-enoyl]oxy}(hydroxy)phosphoryl]adenosine, GLYCEROL, ... | | Authors: | Li, Z, Nair, S.K. | | Deposit date: | 2015-06-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Specificity and Flexibility in a Plant 4-Coumarate:CoA Ligase.
Structure, 23, 2015
|
|
6C9T
 
 | | Transcriptional repressor, CouR | | Descriptor: | CouR | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C2S
 
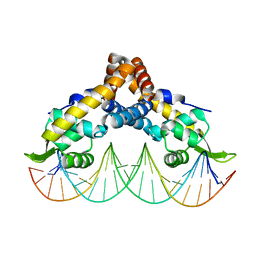 | | Transcriptional repressor, CouR, bound to a 23-mer DNA duplex | | Descriptor: | 23-mer, Transcriptional regulator, MarR family | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6CGQ
 
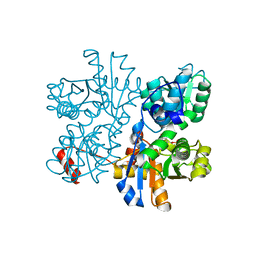 | | Threonine synthase from Bacillus subtilis ATCC 6633 with PLP and PLP-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.
ACS Chem. Biol., 14, 2019
|
|
4MGQ
 
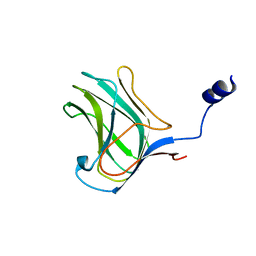 | | PbXyn10C CBM APO | | Descriptor: | CALCIUM ION, Glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6C0Y
 
 | |
4MGS
 
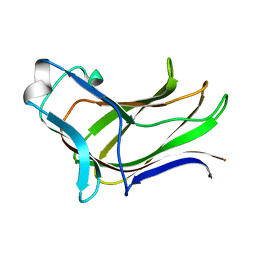 | | BiXyn10A CBM1 APO | | Descriptor: | Putative glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4K8G
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G) | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Lukk, T, Wichelecki, D, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G)
To be Published
|
|
5W8C
 
 | | The structure of a COA-dependent acyl-homoserine lactone synthase, BjaI, with MTA and isovaleryl-CoA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Autoinducer synthase, CHLORIDE ION, ... | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2017-06-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the substrate specificity of quorum signal synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W8G
 
 | |
4ETW
 
 | | Structure of the Enzyme-ACP Substrate Gatekeeper Complex Required for Biotin Synthesis | | Descriptor: | Acyl carrier protein, Pimelyl-[acyl-carrier protein] methyl ester esterase, methyl 7-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxoheptanoate | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5W8A
 
 | |
5UAO
 
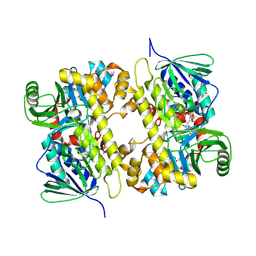 | | Crystal structure of MibH, a lathipeptide tryptophan 5-halogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophane-5-halogenase | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U57
 
 | | Psf4 in complex with Fe2+ and (S)-2-HPP | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, FE (III) ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
2NUM
 
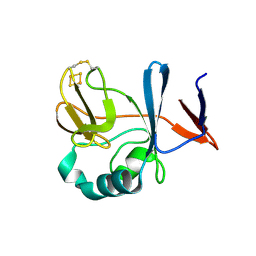 | | Soluble domain of Rieske Iron-Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
5UW6
 
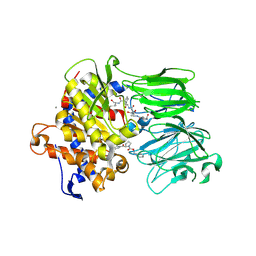 | | PCY1 in Complex with Follower Peptide and Covalent Inhibitor ZPP | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Peptide cyclase 1, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W8D
 
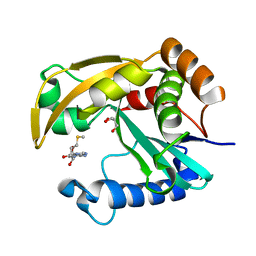 | |
2NUK
 
 | | Soluble Domain of the Rieske Iron-Sulfur Protein from Rhodobacter sphaeroides | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVF
 
 | | Soluble domain of Rieske Iron-Sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NWF
 
 | | Soluble domain of Rieske Iron Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
