1V1O
 
 | |
6XP8
 
 | |
4YAR
 
 | | 2-Hydroxyethylphosphonate dioxygenase (HEPD) E176H | | Descriptor: | 2-hydroxyethylphosphonate dioxygenase, ACETATE ION, CADMIUM ION, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Common Late-Stage Intermediate in Catalysis by 2-Hydroxyethyl-phosphonate Dioxygenase and Methylphosphonate Synthase.
J.Am.Chem.Soc., 137, 2015
|
|
8F72
 
 | | Phage P32 gp64- RNA polymerase | | Descriptor: | MAGNESIUM ION, TPR_REGION domain-containing protein | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-17 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Thermophage RNA polymerase
To Be Published
|
|
6WP9
 
 | | AvaR1 bound to Avenolide | | Descriptor: | (5S)-5-[(6R)-6-hydroxy-6-methyl-5-oxooctyl]furan-2(5H)-one, AvaR1 | | Authors: | Kapoor, I, Olivares, P.J, Nair, S.K. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Elife, 9, 2020
|
|
5F2O
 
 | |
5F2N
 
 | |
8UZD
 
 | |
8CZL
 
 | | Human LanCL1 bound to methyl glutathione (MeGSH) | | Descriptor: | Glutathione S-transferase LANCL1, L-GAMMA-GLUTAMYL-S-METHYLCYSTEINYLGLYCINE, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F5M
 
 | | Crystal structure of P74 gp62 | | Descriptor: | Envelope glycoprotein gp62, MAGNESIUM ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-14 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail.
Nat Commun, 15, 2024
|
|
8D0V
 
 | | Human LanCL1 C264A mutant bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CZK
 
 | |
8D19
 
 | | Human LanCL1 bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5DZT
 
 | |
7QOS
 
 | |
6WPA
 
 | |
5F2K
 
 | |
5EHK
 
 | |
6WP7
 
 | |
2ZEX
 
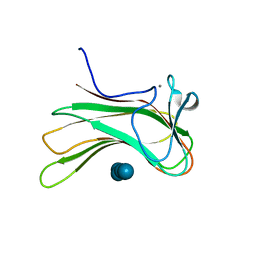 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
2ZEZ
 
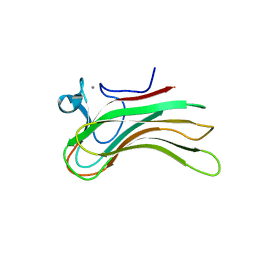 | | Family 16 Carbohydrate Binding Module-2 | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
7RC3
 
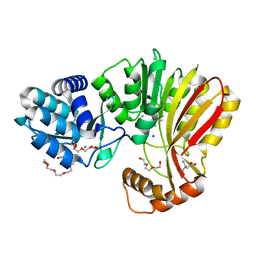 | | Aeronamide N-methyltransferase, AerE (Y137F) | | Descriptor: | ASPARTIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC4
 
 | | Aeronamide N-methyltransferase, AerE (D141A) | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Methyltransferase family protein, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC6
 
 | | Aeronamide N-methyltransferase, AerE, bound to modified peptide substrate, AerA-DL,34 | | Descriptor: | Aeronamide A peptide, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC5
 
 | | Aeronamide N-methyltransferase, AerE (N231A) | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
