1OKN
 
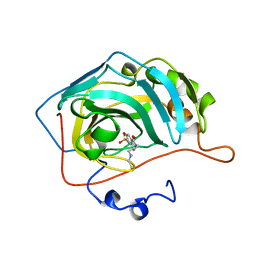 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
6WPA
 
 | |
5DM0
 
 | | Crystal structure of the plantazolicin methyltransferase BamL in complex with triazolic desmethylPZN analog and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ethyl 2-(2-{2-[(1S)-1-amino-4-carbamimidamidobutyl]-1,3-thiazol-4-yl}-5-methyl-1,3-oxazol-4-yl)-1,3-thiazole-4-carboxylate, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
6E6T
 
 | | Dieckmann cyclase, NcmC, bound to cerulenin | | Descriptor: | (4S,5R)-4,5-dihydroxy-5-[(3E,6E)-octa-3,6-dien-1-yl]pyrrolidin-2-one, NcmC, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Enzymatic Off-Loading of Hybrid Polyketides by Dieckmann Condensation.
Acs Chem.Biol., 2020
|
|
6E6U
 
 | | Variant C89S of Dieckmann cyclase, NcmC | | Descriptor: | Dieckmann cyclase, NcmC, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Enzymatic Off-Loading of Hybrid Polyketides by Dieckmann Condensation.
Acs Chem.Biol., 2020
|
|
5DZT
 
 | |
5DM4
 
 | | Crystal structure of the plantazolicin methyltransferase BpumL in complex with pentazolic desmethylPZN analog and SAH | | Descriptor: | 1-[(4S)-4-(4-{4-[4-(5,5'-dimethyl-2,4'-bi-1,3-oxazol-2'-yl)-1,3-thiazol-2-yl]-5-methyl-1,3-oxazol-2-yl}-1,3-thiazol-2-yl)-4-(methylamino)butyl]guanidine, GLYCEROL, Methyltransferase domain family, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
5DM2
 
 | |
6EC7
 
 | |
6EC8
 
 | |
8UZD
 
 | |
5EHK
 
 | |
6WP7
 
 | |
6M7Y
 
 | |
3UQ7
 
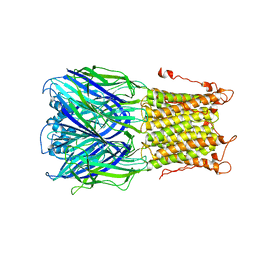 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240S F247L (L9S F16L) in presence of 10 mM cysteamine | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ZEX
 
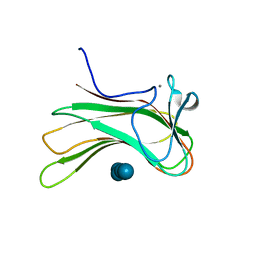 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
2ZEZ
 
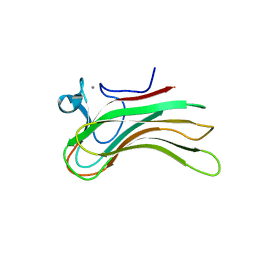 | | Family 16 Carbohydrate Binding Module-2 | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
8T19
 
 | | RiPP precursor peptide recognition element (RRE) domain of Ocin-ThiF-like partner protein, PbtF, bound to an 8 residue fragment of its precursor peptide, PbtA | | Descriptor: | MAGNESIUM ION, PbtA, PbtF | | Authors: | Cogan, D.P, Nair, S.K, Mitchell, D.A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Discovery and validation of RRE domains
To Be Published
|
|
2ZEW
 
 | | Family 16 Cabohydrate Binding Domain Module 1 | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
4E5N
 
 | | Thermostable phosphite dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Thermostable phosphite dehydrogenase | | Authors: | Zou, Y, Zhang, H, Nair, S.K. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of phosphite dehydrogenase provide insights into nicotinamide cofactor regeneration.
Biochemistry, 51, 2012
|
|
3CHH
 
 | | Crystal Structure of Di-iron AurF | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CHI
 
 | |
4E5P
 
 | |
4E5K
 
 | |
4E5M
 
 | |
