6DXX
 
 | | Human N-acylethanolamine-hydrolyzing acid amidase (NAAA) in complex with non-covalent benzothiazole-piperazine inhibitor ARN19702, in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DXY
 
 | | Murine N-acylethanolamine-hydrolyzing acid amidase (NAAA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HQN
 
 | | Catalytic domain of murine Acid Sphingomyelinase (ASMase, ASM, SMPD1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
1MUQ
 
 | | X-ray Crystal Structure of Rattlesnake Venom Complexed With Thiodigalactoside | | Descriptor: | 1-thio-beta-D-galactopyranose-(1-1)-beta-D-galactopyranose, CALCIUM ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2002-09-24 | | Release date: | 2003-07-01 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
5FC7
 
 | | Murine SMPDL3A in complex with sulfate (tetragonal) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
6DY2
 
 | | Guinea pig N-acylethanolamine-hydrolyzing acid amidase (NAAA) covalently bound to beta-lactam inhibitor ARN726 | | Descriptor: | (2S)-3-amino-2-{[(4-cyclohexylbutoxy)carbonyl]amino}propanethioic S-acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DY0
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) covalently bound to beta-lactam inhibitor ARN726, in presence of Triton X-100 | | Descriptor: | (2S)-3-amino-2-{[(4-cyclohexylbutoxy)carbonyl]amino}propanethioic S-acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.014 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DXZ
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) in complex with non-covalent benzothiazole-piperazine inhibitor ARN19702, in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-(2,4,4-trimethylpentan-2-yl)phenol, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DY1
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) with fatty acid (myristate), in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1NVV
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the
Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NVW
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
1NVX
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
4G0M
 
 | | Crystal structure of Arabidopsis thaliana AGO2 MID domain | | Descriptor: | Protein argonaute 2, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
1NVU
 
 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
6DEU
 
 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
6DEV
 
 | | Human caspase-6 E35K | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
4G0O
 
 | | Crystal structure of Arabidopsis thaliana AGO5 MID domain | | Descriptor: | Protein argonaute 5, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0X
 
 | | Crystal Structure of Arabidopsis thaliana AGO1 MID domain | | Descriptor: | Protein argonaute 1, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0P
 
 | | Crystal Structure of Arabidopsis thaliana AGO1 MID domain in complex with UMP | | Descriptor: | Protein argonaute 1, URIDINE-5'-MONOPHOSPHATE | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Y
 
 | | Crystal structure of Arabidopsis AGO1 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Z
 
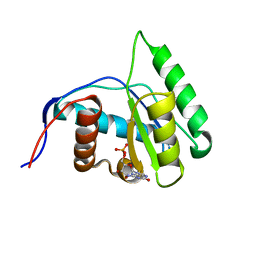 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Q
 
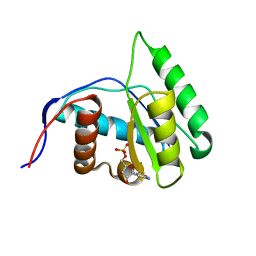 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4K95
 
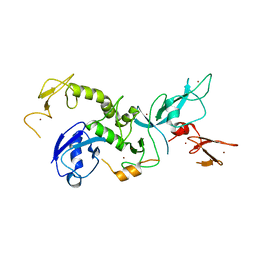 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4HOU
 
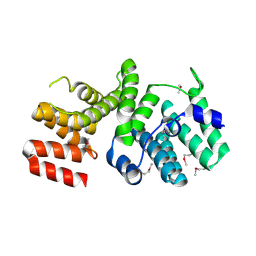 | | Crystal Structure of N-terminal Human IFIT1 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1 | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4IUL
 
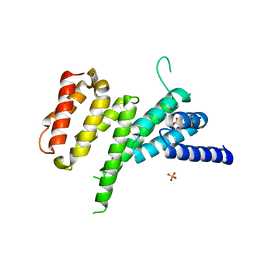 | | MIF4G domain of DAP5 | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 2, SULFATE ION | | Authors: | Frank, F, Virgili, G, Feoktistova, K, Sawicki, M, Sonenberg, N, Fraser, C, Nagar, B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the DAP5 MIF4G Domain and Its Interaction with eIF4A.
Structure, 21, 2013
|
|
