4CWF
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-propyl[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4CWT
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-{[(2Z)-5-(1,3-benzodioxol-5-ylmethyl)-8-fluoro-2-imino-2,3-dihydro[1,2,4]triazolo[1,5-c]quinazolin-10-yl]amino}ethanol, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4CWQ
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-amino-5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazoline-8-sulfonamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
1B0W
 
 | |
6JTA
 
 | | Crystal Structure of D464A L465A mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
1A8T
 
 | | METALLO-BETA-LACTAMASE IN COMPLEX WITH L-159,061 | | Descriptor: | 2-BUTYL-6-HYDROXY-3-[2'-(1H-TETRAZOL-5-YL)-BIPHENYL-4-YLMETHYL]-3H-QUINAZOLIN-4-ONE, METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Fitzgerald, P.M.D, Toney, J.H, Grover, N, Vanderwall, D. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase.
Chem.Biol., 5, 1998
|
|
4D4J
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-bromophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4CKV
 
 | | Crystal structure of VEGFR-1 domain 2 in presence of Zn | | Descriptor: | 1,2-ETHANEDIOL, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1, ZINC ION | | Authors: | Gaucher, J.-F, Reille-Seroussi, M, Gagey-Eilstein, N, Broussy, S, Coric, P, Seijo, B, Lascombe, M.-B, Gautier, B, Liu, W.-Q, Huguenot, F, Inguimbert, N, Bouaziz, S, Vidal, M, Broutin, I. | | Deposit date: | 2014-01-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Plos One, 11, 2016
|
|
6LB5
 
 | | Crystal structure of dimeric RXR-LBD complexed with full agonist NEt-3IB and TIF2 co-activator | | Descriptor: | 6-[ethyl-[3-(2-methylpropoxy)-4-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the full and partial agonist activities of retinoid X receptor alpha ligands with an iso-butoxy and an isopropyl group
Biochem.Biophys.Res.Commun., 734, 2024
|
|
6ICH
 
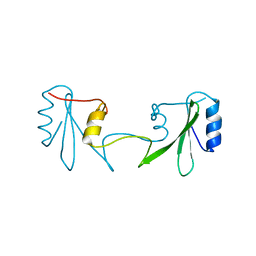 | | Grb2 SH2 domain in domain swapped dimer form | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
6IES
 
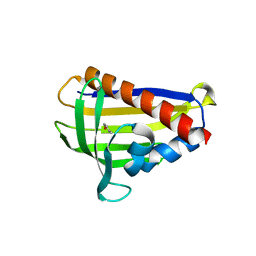 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
2MK0
 
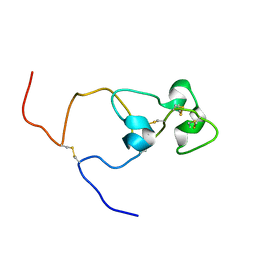 | | Structure of the PSCD4-domain of the cell wall protein pleuralin-1 from the diatom Cylindrotheca fusiformis | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Deutzmann, R, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, S.H.R. | | Deposit date: | 2014-01-22 | | Release date: | 2015-02-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
1B1Y
 
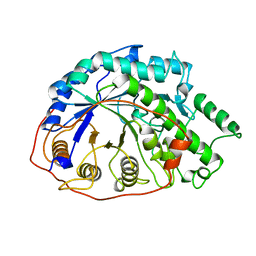 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | Descriptor: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | Authors: | Mikami, B, Yoon, H.J, Yoshigi, N. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
1D2M
 
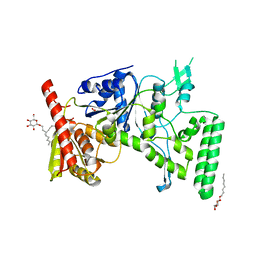 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1CWY
 
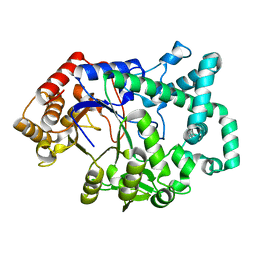 | | CRYSTAL STRUCTURE OF AMYLOMALTASE FROM THERMUS AQUATICUS, A GLYCOSYLTRANSFERASE CATALYSING THE PRODUCTION OF LARGE CYCLIC GLUCANS | | Descriptor: | AMYLOMALTASE | | Authors: | Przylas, I, Tomoo, K, Terada, Y, Takaha, T, Fuji, K, Saenger, W, Straeter, N. | | Deposit date: | 1999-08-27 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amylomaltase from thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans.
J.Mol.Biol., 296, 2000
|
|
1X52
 
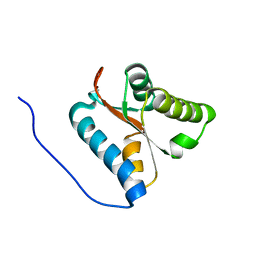 | | Solution structures of the C-terminal domain of the human Pelota homolog (CGI-17) | | Descriptor: | Pelota homolog | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C-terminal domain of the human Pelota homolog (CGI-17)
To be Published
|
|
2PET
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2. | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PF6
 
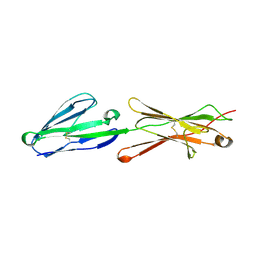 | | Lutheran glycoprotein, N-terminal domains 1 and 2 | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
4V8B
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-06 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8T
 
 | | Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 | | Descriptor: | 25S RIBOSOMAL RNA, 5.8S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Montellese, C, Ban, N. | | Deposit date: | 2012-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-Em Structures of Arx1 and Maturation Factors Rei1 and Jjj1 Bound to the 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 19, 2012
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
7BQQ
 
 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Gogy | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Chantal | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
4W8F
 
 | | Crystal structure of the dynein motor domain in the AMPPNP-bound state | | Descriptor: | Dynein heavy chain lysozyme chimera, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cheng, H.-C, Bhabha, G, Zhang, N, Vale, R.D. | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Allosteric communication in the Dynein motor domain.
Cell, 159, 2014
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Rei | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
