3ZSW
 
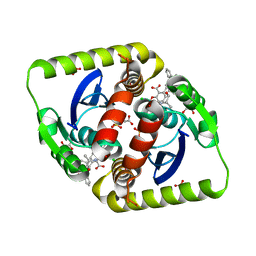 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-[2-[[(2R)-butan-2-yl]carbamoyl]phenyl]methyl-[[(2S)-5-carboxy-2-(2-carboxyethyl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl]-prop-2-enyl-azanium, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3BRE
 
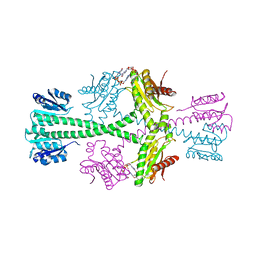 | | Crystal Structure of P.aeruginosa PA3702 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Probable two-component response regulator | | Authors: | De, N, Pirruccello, M, Krasteva, P.V, Bae, N, Raghavan, R.V, Sondermann, H. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation-independent regulation of the diguanylate cyclase WspR.
Plos Biol., 6, 2008
|
|
1HPU
 
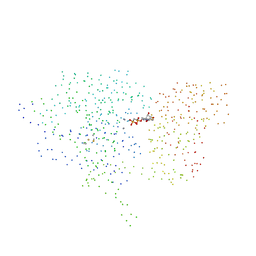 | | 5'-NUCLEOTIDASE (CLOSED FORM), COMPLEX WITH AMPCP | | Descriptor: | 5'-NUCLEOTIDASE, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-13 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
8DPR
 
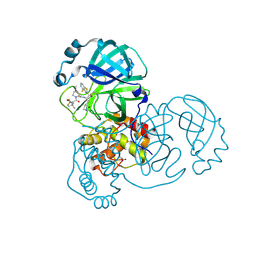 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3ZSR
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT4
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(2-OXO-2,3-DIHYDRO-1H-INDEN-1-YLIDENE)METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design
Plos One, 7, 2012
|
|
3ZSQ
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 5-[[[2-[[(1S)-1-(4-METHOXYPHENYL)BUTYL]CARBAMOYL]PHENYL]METHYL-PROP-2-ENYL-AMINO]METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT3
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-{(E)-[(2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLIDENE]METHYL}-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT0
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (4-CARBOXY-1,3-BENZODIOXOL-5-YL)-N-{2-[(4-METHOXYBENZYL)CARBAMOYL]BENZYL}-N-METHYLMETHANAMINIUM, INTEGRASE, SULFATE ION | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3C7T
 
 | | Crystal structure of the ecdysone phosphate phosphatase, EPPase, from Bombix mori in complex with tungstate | | Descriptor: | CHLORIDE ION, Ecdysteroid-phosphate phosphatase, IODIDE ION, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-02-08 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional characterization of the c-terminal domain of the ecdysteroid phosphate phosphatase from Bombyx mori reveals a new enzymatic activity.
Biochemistry, 47, 2008
|
|
1HP1
 
 | | 5'-NUCLEOTIDASE (OPEN FORM) COMPLEX WITH ATP | | Descriptor: | 5'-NUCLEOTIDASE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-12 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
6EI6
 
 | | CC2D1B coordinates ESRCT-III activity during the mitotic reformation of the nuclear envelope | | Descriptor: | Coiled-coil and C2 domain-containing protein 1-like, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Ventimiglia, L.N, Cuesta-Geijo, M.A, Martinelli, N, Caballe, A, Macheboeuf, P, Miguet, N, Parnham, I.M, Olmos, Y, Carlton, J.G, Weissehorn, W, martin-Serrano, J. | | Deposit date: | 2017-09-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope.
Dev. Cell, 47, 2018
|
|
5WQU
 
 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
2PPS
 
 | | PHOTOSYNTHETIC REACTION CENTER AND CORE ANTENNA SYSTEM (TRIMERIC), ALPHA CARBON ONLY | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHOTOSYSTEM I, ... | | Authors: | Krauss, N, Schubert, W.-D, Klukas, O, Fromme, P, Witt, H.T, Saenger, W. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Photosystem I at 4 A resolution represents the first structural model of a joint photosynthetic reaction centre and core antenna system.
Nat.Struct.Biol., 3, 1996
|
|
5WQS
 
 | | Crystal structure of Apo Beta-Amylase from Sweet potato | | Descriptor: | Beta-amylase, ISOPROPYL ALCOHOL | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
8YTN
 
 | | Single-chain Fv antibody of E11 | | Descriptor: | GLYCEROL, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTO
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTP
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine under reducing conditions | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
2A69
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
3ZSO
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based design | | Descriptor: | 5-{[(2-{[bis(4-methoxyphenyl)methyl]carbamoyl}benzyl)(prop-2-en-1-yl)amino]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Deadman, J.J, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
1HFR
 
 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
4UX6
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
