6S9V
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Thermoanaerobacter thermosaccharolyticum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
6SAM
 
 | | Structure of human butyrylcholinesterase in complex with 1-(2,3-dihydro-1H-inden2-yl)piperidin-3-yl N-phenyl carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Kosak, U, Strasek, N, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2019-07-17 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | N-alkylpiperidine carbamates as potential anti-Alzheimer's agents.
Eur.J.Med.Chem., 197, 2020
|
|
6J1F
 
 | | Crystal structure of HypX from Aquifex aeolicus in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6J3H
 
 | | Crystal structure of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
8KDA
 
 | | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, MAGNESIUM ION, RNA-free ribonuclease P | | Authors: | Teramoto, T, Adachi, N, Yokogawa, T, Koyasu, T, Mayanagi, K, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
8KD8
 
 | |
6J8Y
 
 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
6J81
 
 | | Crystal structure of CERT START domain in complex with compound E14 | | Descriptor: | 2-[4-[4-propyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
8KCV
 
 | | Crystal structure of UDA01-CAAMDDFQL | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of duck MHC UDA01
To Be Published
|
|
6J1I
 
 | |
8KD9
 
 | | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Mayanagi, K, Yokogawa, T, Adachi, N, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
6S8K
 
 | | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to two Tubulin Isotypes | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sharma, A, Olieric, N, Steinmetz, M. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to Two Tubulin Isotypes
Chem, 5, 2019
|
|
6SEG
 
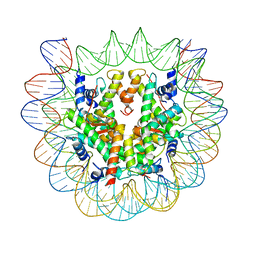 | | Class1: CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
6SG9
 
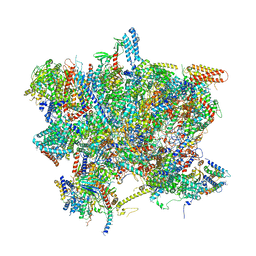 | | Head domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
6SE0
 
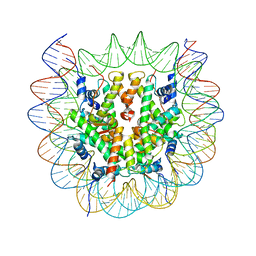 | | Class 1 : CENP-A nucleosome | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
8KGA
 
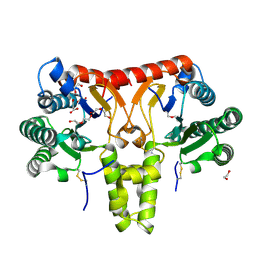 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
6J1J
 
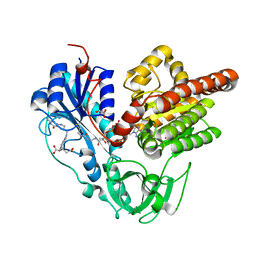 | | Crystal structure of HypX from Aquifex aeolicus, A392F-I419F variant in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
8KGB
 
 | | SlNDPS1-AtcPT4 Chimera complexed with GSPP, Mg2+, and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dimethylallylcistransferase CPT1, chloroplastic,Dehydrodolichyl diphosphate synthase 2, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A versatile system for enzymatic synthesis of natural and unnatural polyisoprenoids on rubber particles
To Be Published
|
|
6SHW
 
 | | N-terminal domain of Drosophila X Virus VP3 | | Descriptor: | SULFATE ION, Structural polyprotein | | Authors: | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | Deposit date: | 2019-08-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
6IXI
 
 | | structure of Cd-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Kumar, P, Sharma, N, Dalal, V, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the heavy metal binding properties of periplasmic metal uptake protein CLas-ZnuA2.
Metallomics, 12, 2020
|
|
6J0J
 
 | |
6J1H
 
 | |
6J3F
 
 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6S8L
 
 | | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to two Tubulin Isotypes | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sharma, A, Olieric, N, Steinmetz, M. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to Two Tubulin Isotypes
Chem, 2019
|
|
