6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NI1
 
 | | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis
To Be Published
|
|
6NPZ
 
 | | Crystal structure of Akt1 (aa 123-480) kinase with a bisubstrate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Cole, P.A, Gabelli, S.B. | | Deposit date: | 2019-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
6NY3
 
 | | CasX ternary complex with 30bp target DNA | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6O2H
 
 | |
6YM5
 
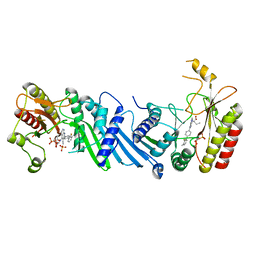 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
4PF3
 
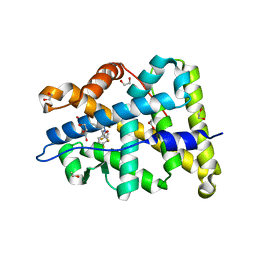 | | Mineralocorticoid receptor ligand-binding domain with compuond 37a | | Descriptor: | 1,2-ETHANEDIOL, 6-[1-(2,2-difluoro-3-hydroxypropyl)-5-(4-fluorophenyl)-3-methyl-1H-pyrazol-4-yl]-2H-1,4-benzoxazin-3(4H)-one, Mineralocorticoid receptor | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2014-04-28 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of 6-[5-(4-fluorophenyl)-3-methyl-pyrazol-4-yl]-benzoxazin-3-one derivatives as novel selective nonsteroidal mineralocorticoid receptor antagonists
Bioorg.Med.Chem., 22, 2014
|
|
6YM3
 
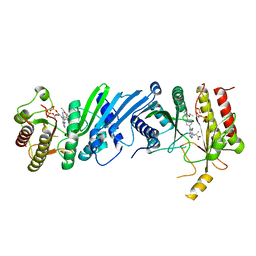 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
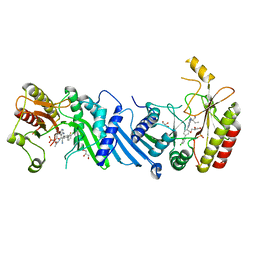 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NX4
 
 | | Structure of the C-terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase (eEF-2K) | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Giles, D.H, Hajredini, F, Dalby, K.N, Ghose, R. | | Deposit date: | 2019-02-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxy-Terminal Tandem Repeat Domain of Eukaryotic Elongation Factor 2 Kinase and Its Role in Substrate Recognition.
J.Mol.Biol., 431, 2019
|
|
6NY2
 
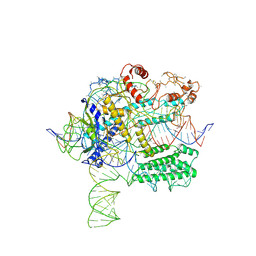 | | CasX-gRNA-DNA(45bp) state I | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
7F1D
 
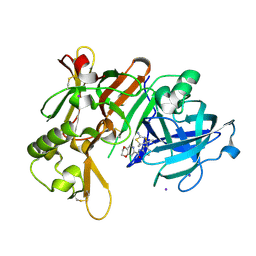 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 1, IODIDE ION, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
7D5A
 
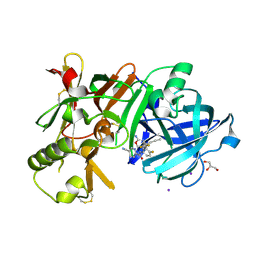 | | Crystal Structure of BACE1 in complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D2X
 
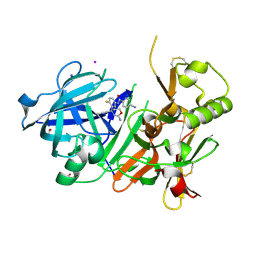 | | Crystal Structure of BACE1 in complex with N-{3-[(4R)-2-amino-4-(prop-1-yn-1-yl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
6NFU
 
 | | Structure of the KcsA-G77A mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NJG
 
 | |
6YEL
 
 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
7BV3
 
 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
6YCD
 
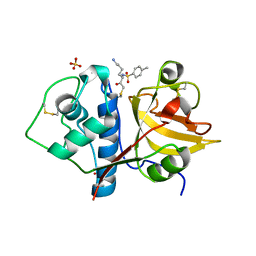 | | Structure the ananain protease from Ananas comosus covalently bound to the TLCK inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
7DCZ
 
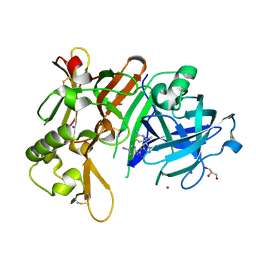 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
6NZ5
 
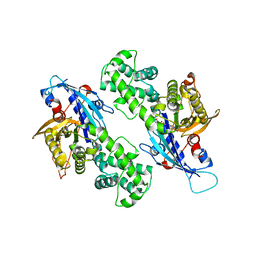 | | YcjX-GDPCP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T, Sung, N, Lee, J, Chang, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6YCC
 
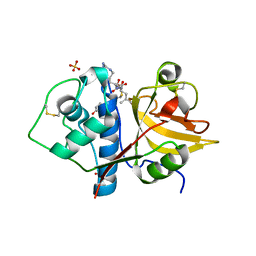 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6O0C
 
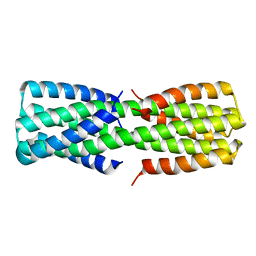 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NUG
 
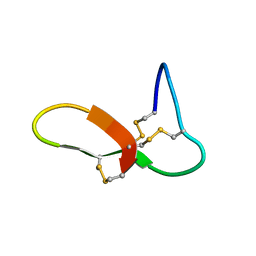 | | hGRNA4-28_3s | | Descriptor: | Granulin-4 | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | hGRN4-28_3s
to be published
|
|
