6VZV
 
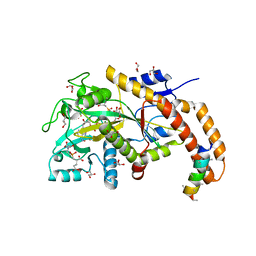 | | TTLL6 bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W1U
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GVI
 
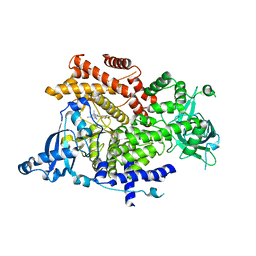 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidine-4,6-diamine | | Descriptor: | 3-(2-azanyl-1,3-benzoxazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-d]pyrimidine-4,6-diamine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6W1T
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PUB
 
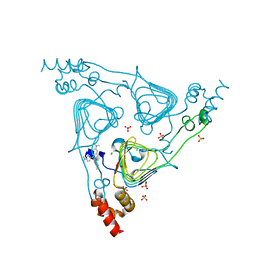 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | Descriptor: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
1SG3
 
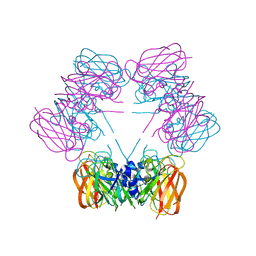 | | Structure of allantoicase | | Descriptor: | Allantoicase | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Graille, M, Meyer, P, Liger, D, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-02-23 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Yeast Allantoicase Reveals a Repeated Jelly Roll Motif
J.Biol.Chem., 279, 2004
|
|
6BZU
 
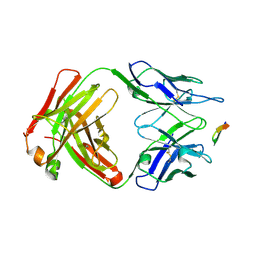 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZY
 
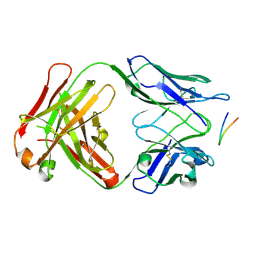 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6UMQ
 
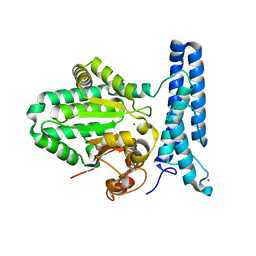 | | Structure of DUF89 | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
6C1E
 
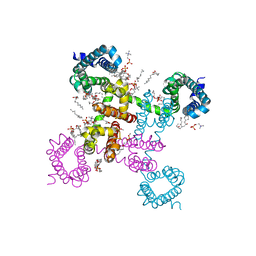 | | NavAb NormoPP mutant | | Descriptor: | (3ALPHA,5ALPHA,7ALPHA,8ALPHA,12ALPHA,14BETA,17ALPHA)-3,7,12-TRIHYDROXYCHOL-1-EN-24-AMIDE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5-BETA-24-NOR-CHOLANE-3(ALPHA),7(ALPHA),12(ALPHA)-TRIOL, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6PXH
 
 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
1SP4
 
 | | Crystal structure of NS-134 in complex with bovine cathepsin B: a two headed epoxysuccinyl inhibitor extends along the whole active site cleft | | Descriptor: | Cathepsin B, methyl N-[(2S)-4-{[(1S)-1-{[(2S)-2-carboxypyrrolidin-1-yl]carbonyl}-3-methylbutyl]amino}-2-hydroxy-4-oxobutanoyl]-L-leucylglycylglycinate | | Authors: | Stern, I, Schaschke, N, Moroder, L, Turk, D. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NS-134 in complex with bovine cathepsin B: a two-headed epoxysuccinyl inhibitor extends along the entire active-site cleft.
Biochem.J., 381, 2004
|
|
6UGM
 
 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6UE9
 
 | | Structure of tetrameric sIgA complex (Class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6O7E
 
 | |
6D1R
 
 | | Structure of Staphylococcus aureus RNase P protein at 2.0 angstrom | | Descriptor: | Ribonuclease P protein component | | Authors: | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of the ribonuclease-P-protein subunit from Staphylococcus aureus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6H00
 
 | | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Pyridoxine-5'-phosphate oxidase, ... | | Authors: | Mackinnon, S, Wilson, M.P, Shrestha, L, Bezerra, G.A, Newman, J, Fox, N, Sorrell, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Clayton, P.T, Mills, P.B, Yue, W.W. | | Deposit date: | 2018-07-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant
To Be Published
|
|
6D2Y
 
 | | Crystal structure of surface glycan-binding protein PbSGBP-B from Prevotella bryantii | | Descriptor: | GLYCEROL, MAGNESIUM ION, PbSGBP-B lipoprotein | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of surface glycan-binding protein PbSGBP-B from Prevotella bryantii
To Be Published
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
5DMK
 
 | | Crystal Structure of IAg7 in complex with RLGL-WE14 | | Descriptor: | CITRATE ANION, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Jin, N, Dai, S, Kappler, J.W. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6O9A
 
 | | Crystal structure of MqnA complexed with 3-hydroxybenzoic acid | | Descriptor: | 3-HYDROXYBENZOIC ACID, ACETATE ION, Chorismate dehydratase | | Authors: | Hicks, K.A, Mahanta, N, Naseem, S, Fedoseyenko, D, Begley, T.P, Ealick, S.E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Menaquinone Biosynthesis: Biochemical and Structural Studies of Chorismate Dehydratase.
Biochemistry, 58, 2019
|
|
6UJI
 
 | |
5DNK
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DPI
 
 | | sfGFP double mutant - 133/149 p-cyano-L-phenylalanine | | Descriptor: | Green fluorescent protein | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-01-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1SDZ
 
 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
