4Y3P
 
 | | Endothiapepsin in complex with fragment 17 | | Descriptor: | 1-(6-methylpyrimidin-4-yl)azepane, Endothiapepsin, GLYCEROL | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y54
 
 | | Endothiapepsin in complex with fragment 56 | | Descriptor: | (2R)-(3-chlorophenyl)(hydroxy)ethanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3E
 
 | | Endothiapepsin in complex with fragment 5 | | Descriptor: | 1H-isoindol-3-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y40
 
 | | Structure of Vaspin mutant D305C V383C | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | Authors: | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
1Q5T
 
 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
4YFB
 
 | |
4YGM
 
 | | Vaccinia virus his-D4/A20(1-50) in complex with uracil | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, URACIL, ... | | Authors: | Tarbouriech, N, Iseni, F, Burmeister, W.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
4Y4U
 
 | | Endothiapepsin in complex with fragment 14 | | Descriptor: | 1-(4-bromo-2-chlorophenyl)-3-methylthiourea, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6N9X
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS3) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
8BEL
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEH
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
4Y6L
 
 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6U
 
 | | Mycobacterial protein | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, CHLORIDE ION, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1PX9
 
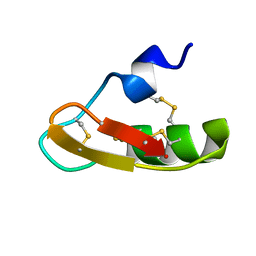 | | Solution structure of the native CnErg1 Ergtoxin, a highly specific inhibitor of HERG channel | | Descriptor: | ergtoxin | | Authors: | Frenal, K, Wecker, K, Gurrola, G.B, Possani, L.D, Wolff, N, Delepierre, M. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Exploring structural features of the interaction between the scorpion toxinCnErg1 and ERG K+ channels.
Proteins, 56, 2004
|
|
8BEF
 
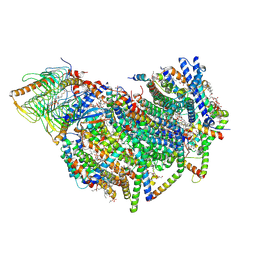 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEE
 
 | |
4YF9
 
 | |
1PZV
 
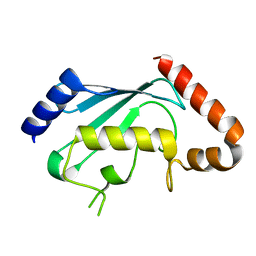 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1Q01
 
 | | Lebetin peptides, a new class of potent aggregation inhibitors | | Descriptor: | lebetin 2 isoform alpha | | Authors: | Mosbah, A, Marrakchi, N, Ganzalez, M.J, Van Rietschoten, J, Giralt, E, El Ayeb, M, Rochat, H, Sabatier, J.M, Darbon, H, Mabrouk, K. | | Deposit date: | 2003-07-15 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Lebetin peptides, a new class of potent aggregation inhibitors
To be Published
|
|
1Q34
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
4YJF
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (S-methylbenzylamine binding form) | | Descriptor: | (1S)-1-phenylethanamine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4Y51
 
 | | Endothiapepsin in complex with fragment 52 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1-(4-bromophenyl)ethanone, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y60
 
 | | Structure of SOX18-HMG/PROX1-DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*AP*GP*CP*AP*TP*TP*GP*TP*CP*TP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*AP*GP*AP*CP*AP*AP*TP*GP*CP*TP*AP*GP*T)-3'), Transcription factor SOX-18 | | Authors: | Narasimhan, K, Prokoph, N, Kolatkar, P, Robinson, H, Jauch, R. | | Deposit date: | 2015-02-12 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction.
Nucleic Acids Res., 44, 2016
|
|
1PUC
 
 | | P13SUC1 IN A STRAND-EXCHANGED DIMER | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, P13SUC1 | | Authors: | Khazanovich, N, Bateman, K.S, Chernaia, M, Michalak, M, James, M.N.G. | | Deposit date: | 1995-12-08 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the yeast cell-cycle control protein, p13suc1, in a strand-exchanged dimer.
Structure, 4, 1996
|
|
1Q3N
 
 | | Crystal structure of KDO8P synthase in its binary complex with substrate PEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-07-31 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
