1MH5
 
 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ8
 
 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1V5V
 
 | |
7X7N
 
 | | 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, Synthetic peptide SIH-5 | | Authors: | Khatri, B, Pramanick, I, Malladi, S.K, Rajmani, R.S, Kumar, S, Ghosh, P, Sengupta, N, Rahisuddin, R, Kumaran, S, Ringe, R.P, Varadarajan, R, Dutta, S, Chatterjee, J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein.
Nat.Chem.Biol., 18, 2022
|
|
5XZF
 
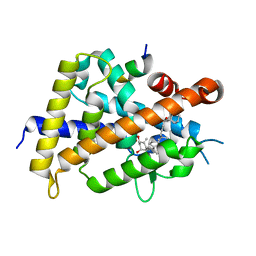 | | Vitamin D receptor with a synthetic ligand ADRO1 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6S)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
7WJL
 
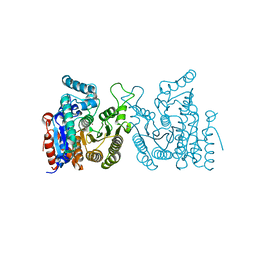 | | Crystal structure of S. cerevisiae Hos3 | | Descriptor: | ACETATE ION, Histone deacetylase HOS3, ZINC ION | | Authors: | Pang, N.N, Che, S.Y, Yang, N. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of fungus-specific histone deacetylase Hos3 provides insights into developing selective inhibitors with antifungal activity.
J.Biol.Chem., 298, 2022
|
|
1Q4N
 
 | |
5XF8
 
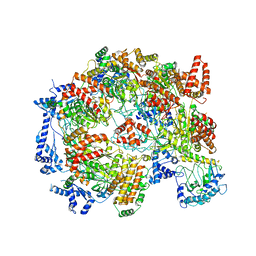 | | Cryo-EM structure of the Cdt1-MCM2-7 complex in AMPPNP state | | Descriptor: | Cell division cycle protein CDT1, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Zhai, Y, Cheng, E, Wu, H, Li, N, Yung, P.Y, Gao, N, Tye, B.K. | | Deposit date: | 2017-04-09 | | Release date: | 2017-05-03 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer
Nat. Struct. Mol. Biol., 24, 2017
|
|
5X7G
 
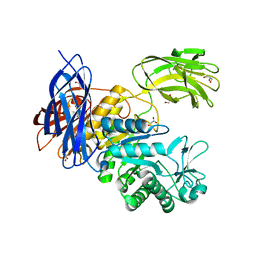 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
7X3K
 
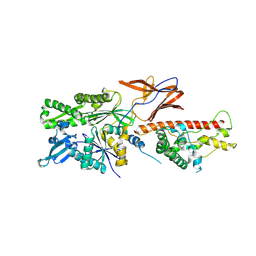 | | Cryo-EM structure of RAC in the State C2 RNC-RAC complex | | Descriptor: | Ribosome-associated complex subunit SSZ1, Zuotin | | Authors: | Chen, Y, Gao, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural remodeling of ribosome associated Hsp40-Hsp70 chaperones during co-translational folding.
Nat Commun, 13, 2022
|
|
7X34
 
 | |
1UL7
 
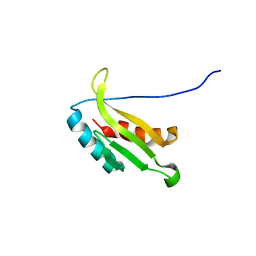 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
6AWB
 
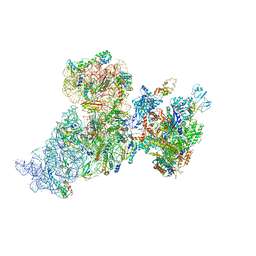 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
1MO6
 
 | |
1MO3
 
 | |
1MJU
 
 | | 1.22 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-12 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PHR
 
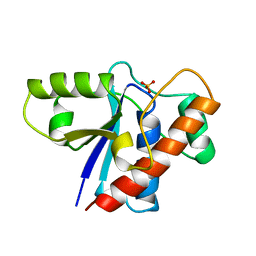 | | THE CRYSTAL STRUCTURE OF A LOW MOLECULAR PHOSPHOTYROSINE PROTEIN PHOSPHATASE | | Descriptor: | LOW MOLECULAR WEIGHT PHOSPHOTYROSINE PROTEIN PHOSPHATASE, SULFATE ION | | Authors: | Su, X.-D, Taddei, N, Stefani, M, Ramponi, G, Nordlund, P. | | Deposit date: | 1994-07-05 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase.
Nature, 370, 1994
|
|
1UB3
 
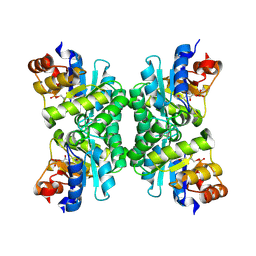 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1QHV
 
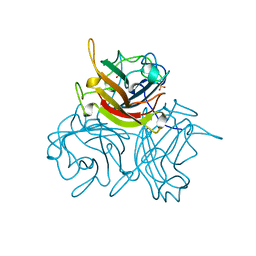 | | HUMAN ADENOVIRUS SEROTYPE 2 FIBRE HEAD | | Descriptor: | PROTEIN (ADENOVIRUS FIBRE), SULFATE ION | | Authors: | Van Raaij, M.J, Louis, N, Chroboczek, J, Cusack, S. | | Deposit date: | 1999-05-28 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the human adenovirus serotype 2 fiber head domain at 1.5 A resolution.
Virology, 262, 1999
|
|
6I7P
 
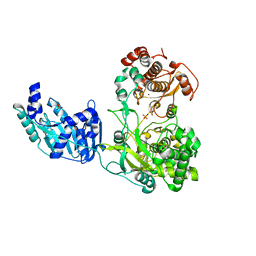 | | Crystal structure of the full-length Zika virus NS5 protein (Human isolate Z1106033) | | Descriptor: | NS5, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Ruiz-Arroyo, V.M, Soler, N, Uson, I, Verdaguer, N. | | Deposit date: | 2018-11-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.975 Å) | | Cite: | Supramolecular arrangement of the full-length Zika virus NS5.
Plos Pathog., 15, 2019
|
|
6IMV
 
 | | The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
6QXZ
 
 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
