8TL9
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
6FSF
 
 | | Crystal structure of the tandem PX-PH-domains of Bem3 from Saccharomyces cerevisiae | | Descriptor: | GTPase-activating protein BEM3 | | Authors: | Ali, I, Eu, S, Koch, D, Bleimling, N, Goody, R.S, Mueller, M.P. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the tandem PX-PH domains of Bem3 from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8TKE
 
 | | Human Type 3 IP3 Receptor - Preactivated+Ca2+ State (+IP3/ATP/JD Ca2+) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
5VRF
 
 | | CryoEM Structure of the Zinc Transporter YiiP from helical crystals | | Descriptor: | Cadmium and zinc efflux pump FieF, ZINC ION | | Authors: | Coudray, N, Lopez-Redondo, M, Zhang, Z, Alexopoulos, J, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2017-05-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FU6
 
 | | Phosphotriesterase PTE_C23_2 | | Descriptor: | FORMIC ACID, POLYACRYLIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
8TKG
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VE3
 
 | | Crystal structure of wild-type persulfide dioxygenase-rhodanese fusion protein from Burkholderia phytofirmans | | Descriptor: | BpPRF, FE (III) ION | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
8TK8
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8TKH
 
 | | Human Type 3 IP3 Receptor - Labile Resting State 1 (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
6FWD
 
 | |
6FMY
 
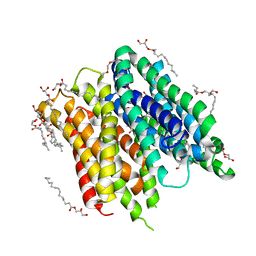 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
5WSF
 
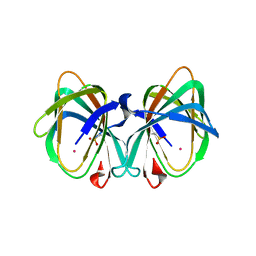 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
6YMN
 
 | |
8THH
 
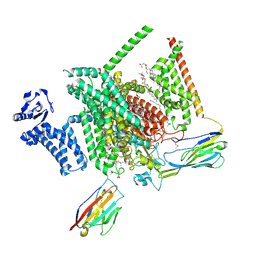 | | Cryo-EM structure of Nav1.7 with LTG | | Descriptor: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6FZG
 
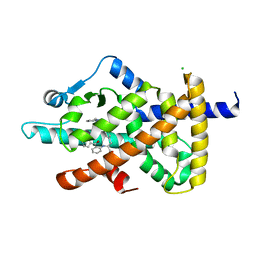 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, Peroxisome proliferator-activated receptor gamma | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
8THG
 
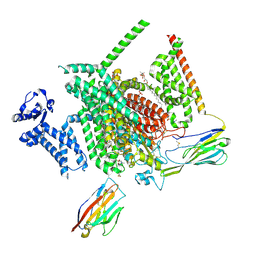 | | Cryo-EM structure of Nav1.7 with RLZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TN3
 
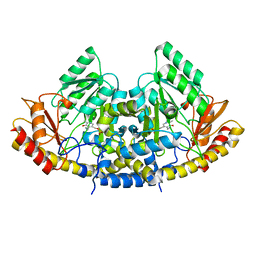 | |
6FRL
 
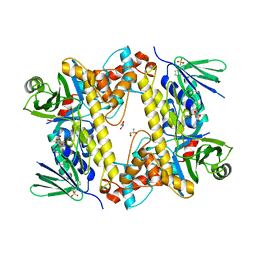 | | BrvH, a flavin-dependent halogenase from Brevundimonas sp. BAL3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Widmann, C, Neubauer, P.R, Sewald, N, Niemann, H.H. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination.
PLoS ONE, 13, 2018
|
|
8TN2
 
 | |
5WUY
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Iqbal, N, Chaudhary, A, Shukla, K.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To Be Published
|
|
6G3M
 
 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
6YFV
 
 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
6YGU
 
 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
8TMT
 
 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
