2G0K
 
 | |
2G0L
 
 | | Solution Structure of Neocarzinostatin Apo-Protein with bound Flavone | | Descriptor: | 2-PHENYL-4H-CHROMEN-4-ONE, NEOCARZINOSTATIN | | Authors: | Muskett, F.W, Stoneman, R.G, Caddick, S, Woolfson, D.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Synthetic Ligands for Apo-Neocarzinostatin
J.Am.Chem.Soc., 128, 2006
|
|
7K4M
 
 | | Crystal structure of MetAP2 Modified Hemoglobin S | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, M.K, Light, D.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MetAP2 inhibition modifies hemoglobin S to delay polymerization and improves blood flow in sickle cell disease.
Blood Adv, 5, 2021
|
|
7JLY
 
 | |
5BYG
 
 | | X-ray structure of AAV2 OBD-AAVS1 complex 2:1 | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Musayev, F.N, Escalante, C.R. | | Deposit date: | 2015-06-10 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Assembly of the Adeno-associated Virus Type 2 Rep68 Protein on the Integration Site AAVS1.
J.Biol.Chem., 290, 2015
|
|
4ZO0
 
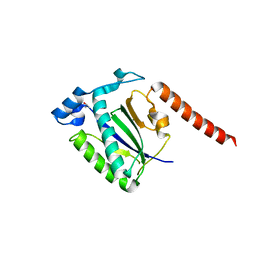 | |
6BWP
 
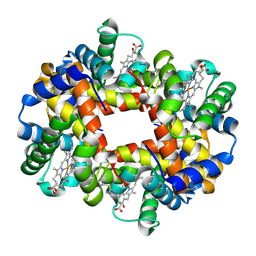 | | Crystal structure of Deoxy Hemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
6BWU
 
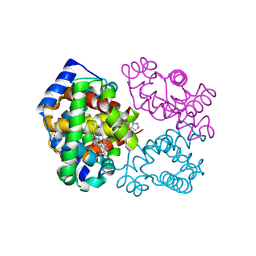 | | Crystal structure of carboxyhemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
5DCX
 
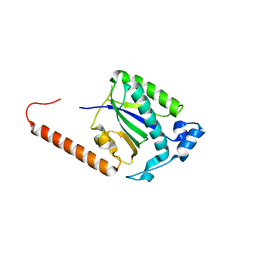 | |
4ZQ9
 
 | | X-ray structure of AAV-2 OBD bound to AAVS1 site 3:1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*CP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*GP*GP*GP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Musayev, F.N, Zarate-Perez, F, Escalante, C.R. | | Deposit date: | 2015-05-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Assembly of the Adeno-associated Virus Type 2 Rep68 Protein on the Integration Site AAVS1.
J.Biol.Chem., 290, 2015
|
|
5V30
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NP6
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5V7S
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E, pH 6.2) | | Descriptor: | Matrix protein 1, PHOSPHATE ION | | Authors: | Musayev, F.N, Safo, M.K, Althufairi, B, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J, Zhou, Q. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V8A
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88R, pH 7.3) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Zhou, Q, Chiang, M.-J, Kosikova, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V6G
 
 | | Crystal structure of Influenza A virus Matrix Protein M1(NLS-88R) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
1PE3
 
 | |
1YLB
 
 | | NMR solution structure of the reduced spinach plastocyanin | | Descriptor: | COPPER (I) ION, Plastocyanin, chloroplast | | Authors: | Musiani, F, Dikiy, A, Semenov, A.Y, Ciurli, S. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intermolecular Complex between Plastocyanin and Cytochrome f from Spinach.
J.Biol.Chem., 280, 2005
|
|
5V7B
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
2TMP
 
 | | N-TERMINAL DOMAIN OF TISSUE INHIBITOR OF METALLOPROTEINASE-2 (N-TIMP-2), NMR, 49 STRUCTURES | | Descriptor: | TISSUE INHIBITOR OF METALLOPROTEINASES-2 | | Authors: | Muskett, F.W, Frenkiel, T.A, Feeney, J, Freedman, R.B, Carr, M.D, Williamson, R.A. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the N-terminal domain of tissue inhibitor of metalloproteinases-2 and characterization of its interaction site with matrix metalloproteinase-3.
J.Biol.Chem., 273, 1998
|
|
1NRG
 
 | | Structure and Properties of Recombinant Human Pyridoxine-5'-Phosphate Oxidase | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Musayev, F.N, di Salvo, M.L, Ko, T.P, Schirch, V, Safo, M.K. | | Deposit date: | 2003-01-24 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and properties of recombinant human pyridoxine 5'-phosphate oxidase.
Protein Sci., 12, 2003
|
|
1U6E
 
 | | 1.85 Angstrom Crystal Structure of the C112A Mutant of Mycobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III, CHLORIDE ION | | Authors: | Mussayev, F, Sachedeva, S, Scarsdale, J.N, Reynolds, K.A, Wright, H.T. | | Deposit date: | 2004-07-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A.
J.Mol.Biol., 346, 2005
|
|
1U6S
 
 | | Crystal Structure of the Complex Between Mycobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III and Lauroyl Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III, DODECYL-COA | | Authors: | Musayev, F, Sachdeva, S, Scarsdale, J.N, Reynolds, K.A, Wright, H.T. | | Deposit date: | 2004-07-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A.
J.Mol.Biol., 346, 2005
|
|
2L1M
 
 | |
1HKO
 
 | | NMR structure of bovine cytochrome b5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muskett, F.W, Whitford, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bovine Ferricytochrome B5 Determined Using Heteronuclear NMR Methods.
J.Mol.Biol., 258, 1996
|
|
5SVB
 
 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
