5AVG
 
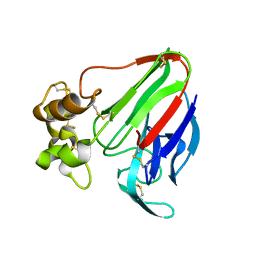 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
3AGV
 
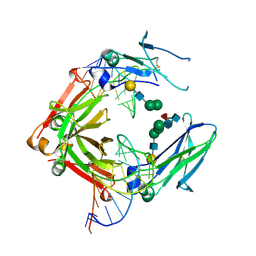 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
4K7Q
 
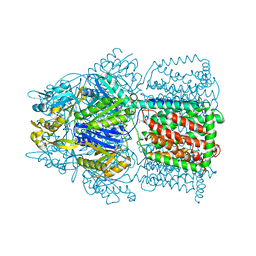 | | Crystal Structure of AcrB Complexed with Linezolid at 3.5 Resolution | | Descriptor: | Acriflavine resistance protein B, N-{[(5S)-3-(3-fluoro-4-morpholin-4-ylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide | | Authors: | Hung, L.W, Kim, H.B, Murakami, S, Gupta, G, Kim, C.Y, Terwilliger, T.C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB complexed with linezolid at 3.5 Angstrom resolution.
J.Struct.Funct.Genom., 14, 2013
|
|
2ZUQ
 
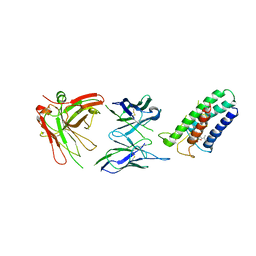 | | Crystal structure of DsbB-Fab complex | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
2ZUP
 
 | | Updated crystal structure of DsbB-DsbA complex from E. coli | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S, Nakagawa, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
