1FIY
 
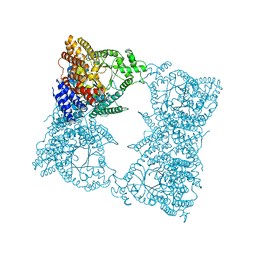 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4YSO
 
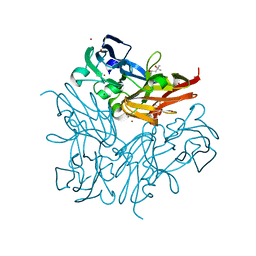 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSA
 
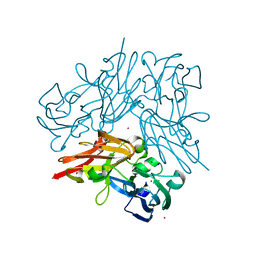 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YST
 
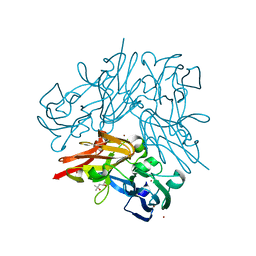 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
7W78
 
 | | Heme exporter HrtBA in complex with Mg-AMPPNP | | Descriptor: | ACETATE ION, DODECANE, GLYCEROL, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YSU
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
1RCK
 
 | |
1R5Z
 
 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7VLE
 
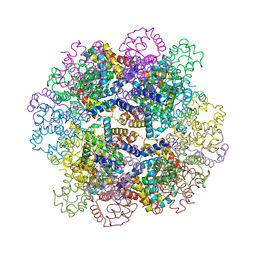 | | Oxy-deoxy intermediate of V2 hemoglobin at 55% oxygen saturation | | Descriptor: | Extracellular A1 globin, Extracellular A2 globin, Extracellular B1 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
1RCL
 
 | |
1SU4
 
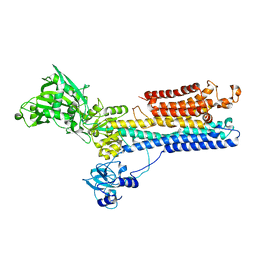 | | Crystal structure of calcium ATPase with two bound calcium ions | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Toyoshima, C, Nakasako, M, Nomura, H, Ogawa, H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution
Nature, 405, 2000
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | Descriptor: | TRF1 | | Authors: | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
6JB8
 
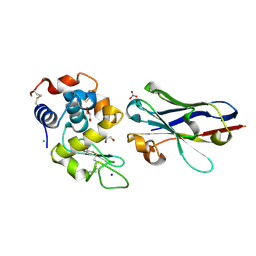 | | Crystal structure of nanobody D3-L11 in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB5
 
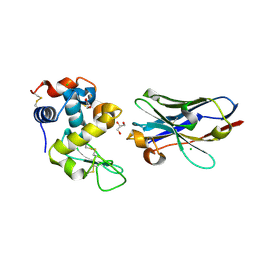 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme (form II) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
1WTJ
 
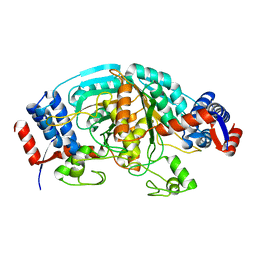 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | Descriptor: | ureidoglycolate dehydrogenase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1WDD
 
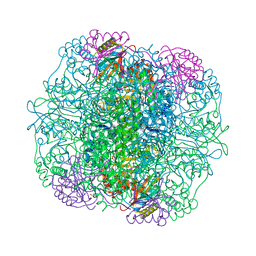 | | Crystal Structure of Activated Rice Rubisco Complexed with 2-Carboxyarabinitol-1,5-bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Ueno, T, Ishida, H, Inoue, T, Makino, A, Mae, T, Kai, Y. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
1WSF
 
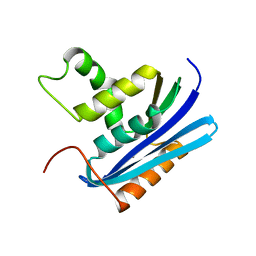 | | Co-crystal structure of E.coli RNase HI active site mutant (D134A*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
1WN7
 
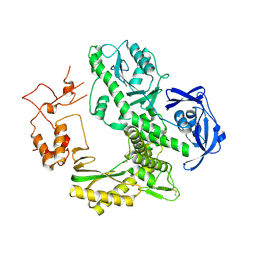 | | Crystal structure of archaeal family B DNA polymerase mutant | | Descriptor: | Family B DNA Polymerase, GLYCEROL, NICKEL (II) ION | | Authors: | Kuroita, T, Matsumura, H, Yokota, N, Hashimoto, H, Imanaka, T, Inoue, T, Kai, Y. | | Deposit date: | 2004-07-28 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Mechanism for Coordination of Proofreading and Polymerase Activities in Archaeal DNA Polymerases
J.Mol.Biol., 351, 2005
|
|
1WTG
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-biphenylalanine-Gln-p-aminobenzamidine | | Descriptor: | 2-(3-BIPHENYL-4-YL-2-ETHANESULFONYLAMINO-PROPIONYLAMINO)-PENTANEDIOIC ACID 5-AMIDE 1-(4-CARBAMIMIDOYL-BENZYLAMIDE), CALCIUM ION, Coagulation factor VII, ... | | Authors: | Kadono, S, Sakamoto, S, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Kitazawa, K, Yoshihashi, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Kodama, M, Haramura, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-23 | | Release date: | 2005-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel interactions of large P3 moiety and small P4 moiety in the binding of the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 326, 2005
|
|
1WSG
 
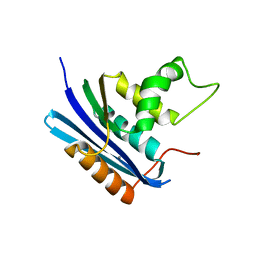 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A/D134N*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
6JB9
 
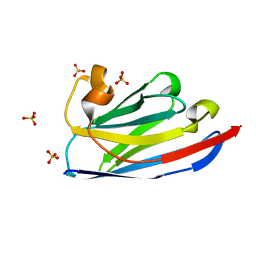 | | Crystal structure of nanobody D3-L11 (unbound form) | | Descriptor: | Nanobody D3-L11, SULFATE ION | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
1IXF
 
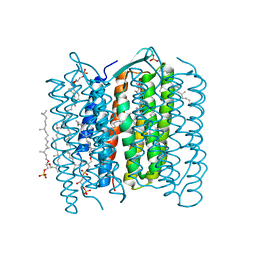 | | Crystal Structure of the K intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Matsui, Y, Sakai, K, Murakami, M, Shiro, Y, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin
J.MOL.BIOL., 324, 2002
|
|
5H5H
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5G
 
 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
1WPG
 
 | | Crystal structure of the SR CA2+-ATPase with MGF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T. | | Deposit date: | 2004-09-02 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
